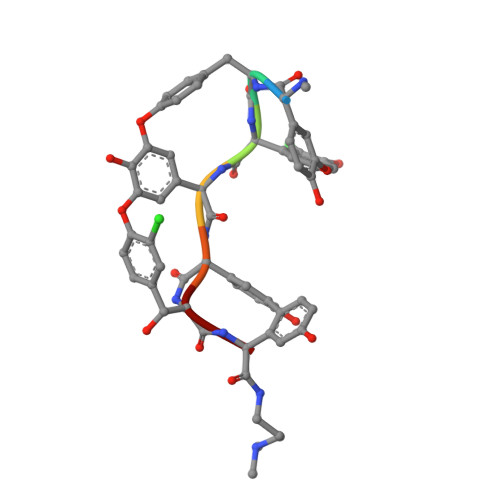
A carrier protein strategy yields the structure of dalbavancin.
Economou, N.J., Nahoum, V., Weeks, S.D., Grasty, K.C., Zentner, I.J., Townsend, T.M., Bhuiya, M.W., Cocklin, S., Loll, P.J.(2012) J Am Chem Soc 134: 4637-4645
- PubMed: 22352468
- DOI: https://doi.org/10.1021/ja208755j
- Primary Citation of Related Structures:
3RUL, 3RUM, 3RUN - PubMed Abstract:
Many large natural product antibiotics act by specifically binding and sequestering target molecules found on bacterial cells. We have developed a new strategy to expedite the structural analysis of such antibiotic-target complexes, in which we covalently link the target molecules to carrier proteins, and then crystallize the entire carrier-target-antibiotic complex. Using native chemical ligation, we have linked the Lys-D-Ala-D-Ala binding epitope for glycopeptide antibiotics to three different carrier proteins. We show that recognition of this peptide by multiple antibiotics is not compromised by the presence of the carrier protein partner, and use this approach to determine the first-ever crystal structure for the new therapeutic dalbavancin. We also report the first crystal structure of an asymmetric ristocetin antibiotic dimer, as well as the structure of vancomycin bound to a carrier-target fusion. The dalbavancin structure reveals an antibiotic molecule that has closed around its binding partner; it also suggests mechanisms by which the drug can enhance its half-life by binding to serum proteins, and be targeted to bacterial membranes. Notably, the carrier protein approach is not limited to peptide ligands such as Lys-D-Ala-D-Ala, but is applicable to a diverse range of targets. This strategy is likely to yield structural insights that accelerate new therapeutic development.
- Department of Biochemistry and Molecular Biology, Drexel University College of Medicine, Philadelphia, Pennsylvania 19102, United States.
Organizational Affiliation: