Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and PD-L2
Lazar-Molnar, E., Ramagopal, U.A., Nathenson, S.G., Almo, S.C.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Programmed cell death 1 ligand 2 | A [auth B] | 201 | Mus musculus | Mutation(s): 0 Gene Names: B7dc, Btdc, Cd273, Murine, Pdcd1lg2, Pdl2 | 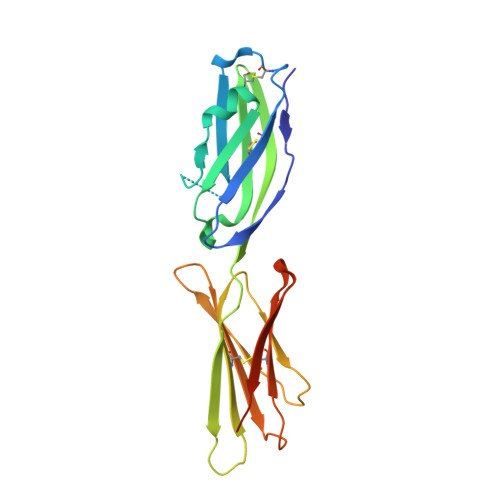 |
UniProt | |||||
Find proteins for Q9WUL5 (Mus musculus) Explore Q9WUL5 Go to UniProtKB: Q9WUL5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9WUL5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Programmed cell death protein 1 | B [auth A] | 117 | Mus musculus | Mutation(s): 2 Gene Names: Pd1, Pdcd1 | 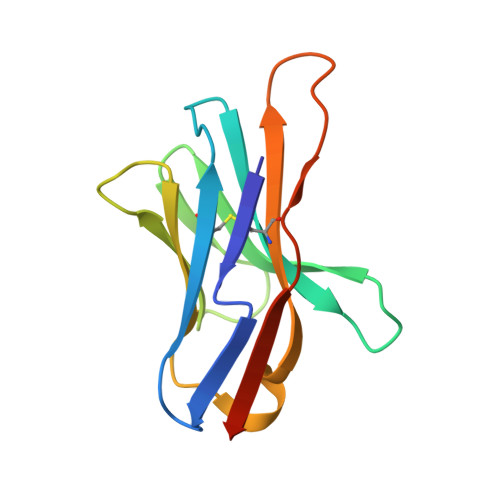 |
UniProt | |||||
Find proteins for Q02242 (Mus musculus) Explore Q02242 Go to UniProtKB: Q02242 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02242 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 36.516 | α = 90 |
| b = 79.716 | β = 105.56 |
| c = 51.693 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| CBASS | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MOLREP | phasing |