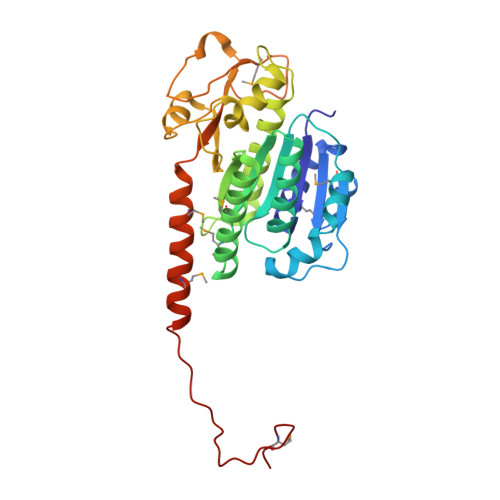
A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell.
Kraemer, J.A., Erb, M.L., Waddling, C.A., Montabana, E.A., Zehr, E.A., Wang, H., Nguyen, K., Pham, D.S., Agard, D.A., Pogliano, J.(2012) Cell 149: 1488-1499
- PubMed: 22726436
- DOI: https://doi.org/10.1016/j.cell.2012.04.034
- Primary Citation of Related Structures:
3R4V, 3RB8 - PubMed Abstract:
Tubulins are essential for the reproduction of many eukaryotic viruses, but historically, bacteriophage were assumed not to require a cytoskeleton. Here, we identify a tubulin-like protein, PhuZ, from bacteriophage 201φ2-1 and show that it forms filaments in vivo and in vitro. The PhuZ structure has a conserved tubulin fold, with an unusual, extended C terminus that we demonstrate to be critical for polymerization in vitro and in vivo. Longitudinal packing in the crystal lattice mimics packing observed by EM of in-vitro-formed filaments, indicating how interactions between the C terminus and the following monomer drive polymerization. PhuZ forms a filamentous array that is required for positioning phage DNA within the bacterial cell. Correct positioning to the cell center and optimal phage reproduction only occur when the PhuZ filament is dynamic. Thus, we show that PhuZ assembles a spindle-like array that functions analogously to the microtubule-based spindles of eukaryotes.
- Department of Biochemistry and Biophysics and the Howard Hughes Medical Institute, University of California, San Francisco, San Francisco, CA 94158, USA.
Organizational Affiliation: