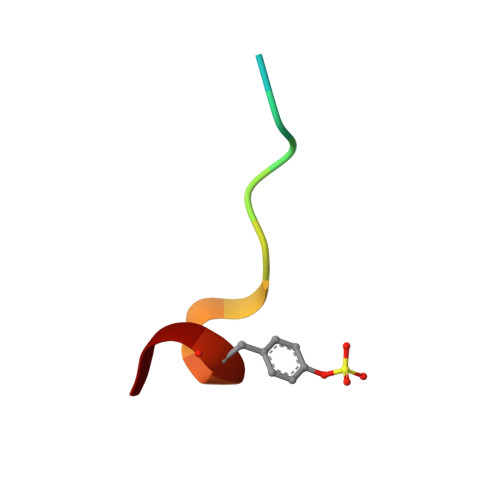
Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
Biela, A., Khayat, M., Tan, H., Kong, J., Heine, A., Hangauer, D., Klebe, G.(2012) J Mol Biology 418: 350-366
- PubMed: 22366545
- DOI: https://doi.org/10.1016/j.jmb.2012.01.054
- Primary Citation of Related Structures:
3P17, 3QTO, 3QTV, 3QWC, 3QX5, 3SHA, 3SHC, 3SI3, 3SI4, 3SV2 - PubMed Abstract:
In the present study, we investigate the impact of a tightly bound water molecule on ligand binding in the S1 pocket of thrombin. The S1 pocket contains a deeply buried deprotonated aspartate residue (Asp189) that is, due to its charged state, well hydrated in the uncomplexed state. We systematically studied the importance of this water molecule by evaluating a series of ligands that contains pyridine-type P1 side chains that could potentially alter the binding properties of this water molecule. All of the pyridine derivatives retain the original hydration state albeit sometimes with a slight perturbance. In order to prevent a direct H-bond formation with Asp189, and to create a permanent positive charge on the P1 side chain that is positioned adjacent to the Asp189 carboxylate anion, we methylated the pyridine nitrogen. This methylation resulted in displacement of water but was accompanied by a loss in binding affinity. Quantum chemical calculations of the ligand solvation free energy showed that the positively charged methylpyridinium derivatives suffer a large penalty of desolvation upon binding. Consequently, they have a substantially less favorable enthalpy of binding. In addition to the ligand desolvation penalty, the hydration shell around Asp189 has to be overcome, which is achieved in nearly all pyridinium derivatives. Only for the ortho derivative is a partial population of a water next to Asp189 found. Possibly, the gain of electrostatic interactions between the charged P1 side chain and Asp189 helps to compensate for the desolvation penalty. In all uncharged pyridine derivatives, the solvation shell remains next to Asp189, partly mediating interactions between ligand and protein. In the case of the para-pyridine derivative, a strongly disordered cluster of water sites is observed between ligand and Asp189.
- Department of Pharmaceutical Chemistry, Philipps University Marburg, Marbacher Weg 6, 35032 Marburg, Germany.
Organizational Affiliation: