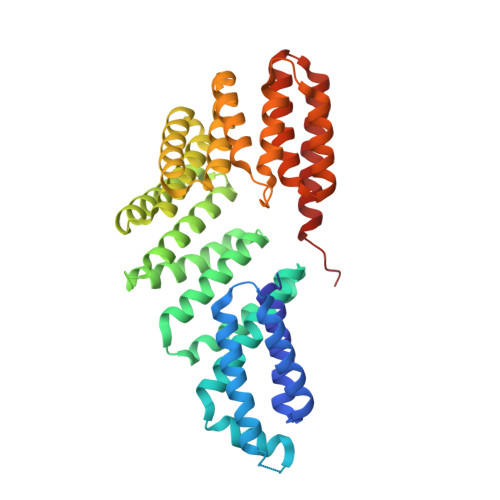
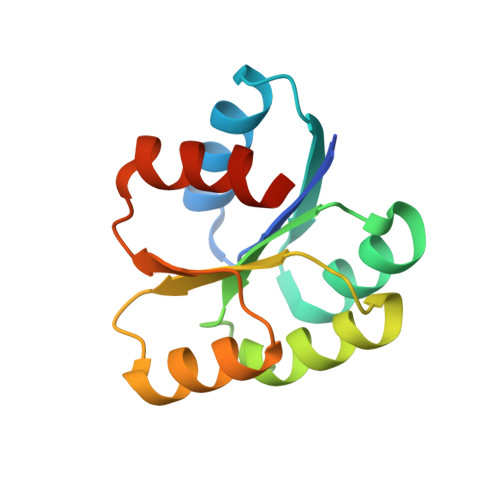
Structural basis of response regulator dephosphorylation by Rap phosphatases.
Parashar, V., Mirouze, N., Dubnau, D.A., Neiditch, M.B.(2011) PLoS Biol 9: e1000589-e1000589
- PubMed: 21346797
- DOI: https://doi.org/10.1371/journal.pbio.1000589
- Primary Citation of Related Structures:
3Q15 - PubMed Abstract:
Bacterial Rap family proteins have been most extensively studied in Bacillus subtilis, where they regulate activities including sporulation, genetic competence, antibiotic expression, and the movement of the ICEBs1 transposon. One subset of Rap proteins consists of phosphatases that control B. subtilis and B. anthracis sporulation by dephosphorylating the response regulator Spo0F. The mechanistic basis of Rap phosphatase activity was unknown. Here we present the RapH-Spo0F X-ray crystal structure, which shows that Rap proteins consist of a 3-helix bundle and a tetratricopeptide repeat domain. Extensive biochemical and genetic functional studies reveal the importance of the observed RapH-Spo0F interactions, including the catalytic role of a glutamine in the RapH 3-helix bundle that inserts into the Spo0F active site. We show that in addition to dephosphorylating Spo0F, RapH can antagonize sporulation by sterically blocking phosphoryl transfer to and from Spo0F. Our structure-function analysis of the RapH-Spo0F interaction identified Rap protein residues critical for Spo0F phosphatase activity. This information enabled us to assign Spo0F phosphatase activity to a Rap protein based on sequence alone, which was not previously possible. Finally, as the ultimate test of our newfound understanding of the structural requirements for Rap phosphatase function, a non-phosphatase Rap protein that inhibits the binding of the response regulator ComA to DNA was rationally engineered to dephosphorylate Spo0F. In addition to revealing the mechanistic basis of response regulator dephosphorylation by Rap proteins, our studies support the previously proposed T-loop-Y allostery model of receiver domain regulation that restricts the aromatic "switch" residue to an internal position when the β4-α4 loop adopts an active-site proximal conformation.
- Department of Microbiology and Molecular Genetics, UMDNJ-New Jersey Medical School, Newark, New Jersey, United States of America.
Organizational Affiliation: