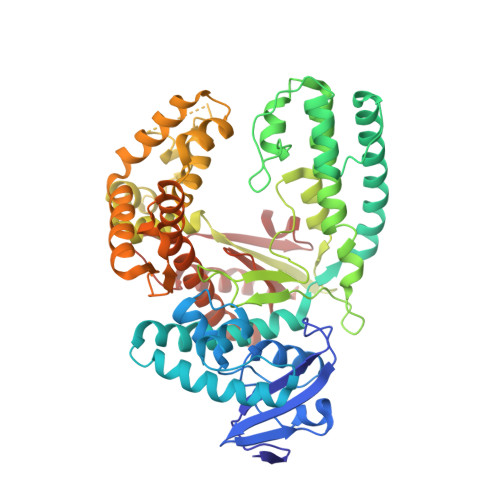
Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Obeid, S., Schnur, A., Gloeckner, C., Blatter, N., Welte, W., Diederichs, K., Marx, A.(2011) Chembiochem 12: 1574-1580
- PubMed: 21480455
- DOI: https://doi.org/10.1002/cbic.201000783
- Primary Citation of Related Structures:
3PO4, 3PO5, 3PY8 - PubMed Abstract:
DNA is being constantly damaged by endo- and exogenous agents such as reactive oxygen species, chemicals, radioactivity, and ultraviolet radiation. Additionally, DNA is inherently labile, and this can result in, for example, the spontaneous hydrolysis of the glycosidic bond that connects the sugar and the nucleobase moieties in DNA; this results in abasic sites. It has long been obscure how cells achieve DNA synthesis past these lesions, and only recently has it been discovered that several specialized DNA polymerases are involved in translesion synthesis. The underlying mechanisms that render one DNA polymerase competent in translesion synthesis while another DNA polymerase fails are still indistinct. Recently two variants of Taq DNA polymerase that exhibited higher lesion bypass ability than the wild-type enzyme were identified by directed-evolution approaches. Strikingly, in both approaches it was independently found that substitution of a single nonpolar amino acid side chain by a cationic side chain increases the capability of translesion synthesis. Here, we combined both mutations in a single enzyme. We found that the KlenTaq DNA polymerase that bore both mutations superseded the wild-type as well as the respective single mutants in translesion-bypass proficiency. Further insights in the molecular basis of the detected gain of translesion-synthesis function were obtained by structural studies of DNA polymerase variants caught in processing canonical and damaged substrates. We found that increased positive charge of the surface potential in the area proximal to the negatively charged substrates promotes translesion synthesis by KlenTaq DNA polymerase, an enzyme that has very limited naturally evolved capability to perform translesion synthesis. Since expanded positively charged surface potential areas are also found in naturally evolved translesion DNA polymerases, our results underscore the impact of charge on the proficiency of naturally evolved translesion DNA polymerases.
- Department of Chemistry, Konstanz Research School Chemical Biology, University of Konstanz, Konstanz, Germany.
Organizational Affiliation: