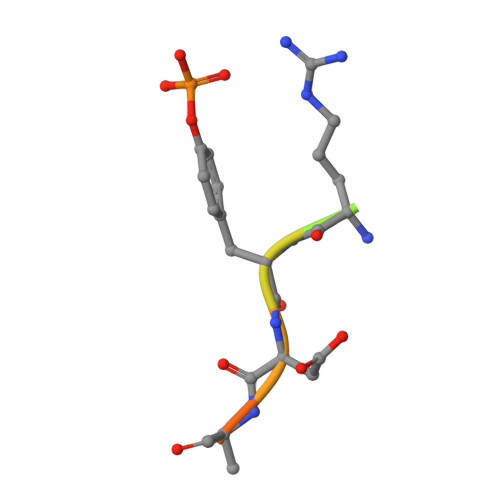
An adjacent arginine, and the phosphorylated tyrosine in the c-Met receptor target sequence, dictates the orientation of c-Cbl binding
Sun, Q., Ng, C., Guy, G.R., Sivaraman, J.(2011) FEBS Lett 585: 281-285
- PubMed: 21163258
- DOI: https://doi.org/10.1016/j.febslet.2010.11.060
- Primary Citation of Related Structures:
3PLF - PubMed Abstract:
Previously, we have demonstrated that the tyrosine phosphorylated hepatocyte growth factor receptor (Met) binds to the c-Cbl phosphotyrosine-recognition, tyrosine kinase binding (TKB) domain in a reverse orientation compared to other c-Cbl binding partners. A Met peptide with the DpYR motif changed to RpYD (MetRD) retains a similar TKB binding affinity as the native Met peptide. However, the TKB: MetRD complex crystal structure reveals a complete reversal of the binding orientation. Collated data indicates that both binding and orientation is dictated by the phosphorylated tyrosine and an adjacent arginine forming intra-peptide hydrogen bonds and aligning unidirectionally with complementary charges in the phosphotyrosine binding pocket of c-Cbl.
- Department of Biological Sciences, National University of Singapore, Singapore, Singapore.
Organizational Affiliation: