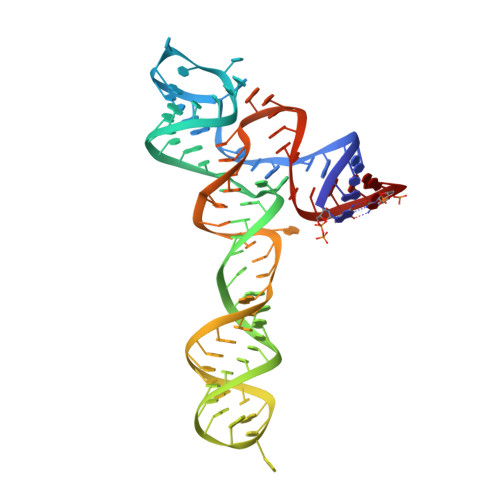
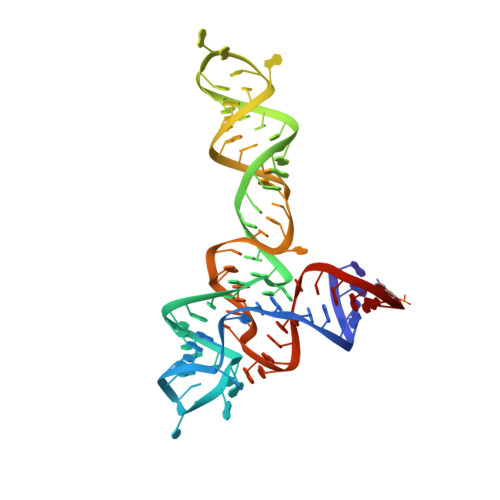
Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Huang, L., Serganov, A., Patel, D.J.(2010) Mol Cell 40: 774-786
- PubMed: 21145485
- DOI: https://doi.org/10.1016/j.molcel.2010.11.026
- Primary Citation of Related Structures:
3OWI, 3OWW, 3OWZ, 3OX0, 3OXB, 3OXD, 3OXE, 3OXJ, 3OXM - PubMed Abstract:
Glycine riboswitches regulate gene expression by feedback modulation in response to cooperative binding to glycine. Here, we report on crystal structures of the second glycine-sensing domain from the Vibrio cholerae riboswitch in the ligand-bound and unbound states. This domain adopts a three-helical fold that centers on a three-way junction and accommodates glycine within a bulge-containing binding pocket above the junction. Glycine recognition is facilitated by a pair of bound Mg(2+) cations and governed by specific interactions and shape complementarity with the pocket. A conserved adenine extrudes from the binding pocket and intercalates into the junction implying that glycine binding in the context of the complete riboswitch could impact on gene expression by stabilizing the riboswitch junction and regulatory P1 helix. Analysis of riboswitch interactions in the crystal and footprinting experiments indicates that adjacent glycine-sensing modules of the riboswitch could form specific interdomain interactions, thereby potentially contributing to the cooperative response.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: