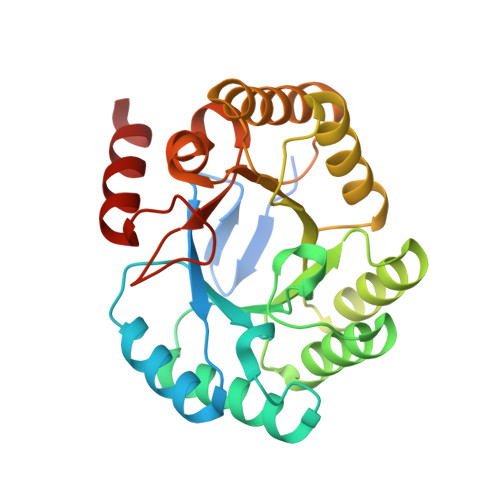
A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding.
Light, S.H., Minasov, G., Shuvalova, L., Peterson, S.N., Caffrey, M., Anderson, W.F., Lavie, A.(2011) Biochemistry 50: 2357-2363
- PubMed: 21291284
- DOI: https://doi.org/10.1021/bi102020s
- Primary Citation of Related Structures:
3L2I, 3O1N, 3OEX - PubMed Abstract:
Dehydroquinate dehydratase (DHQD) catalyzes the third step in the biosynthetic shikimate pathway. We present three crystal structures of the Salmonella enterica type I DHQD that address the functionality of a surface loop that is observed to close over the active site following substrate binding. Two wild-type structures with differing loop conformations and kinetic and structural studies of a mutant provide evidence of both direct and indirect mechanisms of involvement of the loop in substrate binding. In addition to allowing amino acid side chains to establish a direct interaction with the substrate, closure of the loop necessitates a conformational change of a key active site arginine, which in turn positions the substrate productively. The absence of DHQD in humans and its essentiality in many pathogenic bacteria make the enzyme a target for the development of nontoxic antimicrobials. The structures and ligand binding insights presented here may inform the design of novel type I DHQD inhibiting molecules.
- Center for Structural Genomics of Infectious Diseases, Feinberg School of Medicine, Northwestern University, Chicago, Illinois 60611, United States.
Organizational Affiliation: