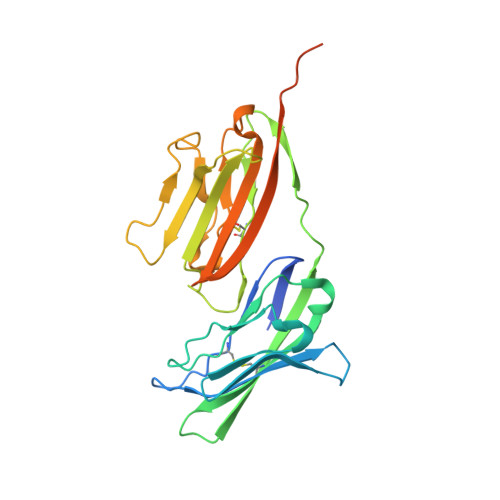
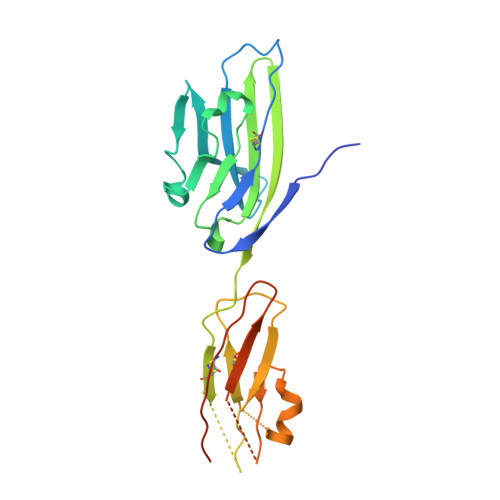
The molecular interaction of CAR and JAML recruits the central cell signal transducer PI3K.
Verdino, P., Witherden, D.A., Havran, W.L., Wilson, I.A.(2010) Science 329: 1210-1214
- PubMed: 20813955
- DOI: https://doi.org/10.1126/science.1187996
- Primary Citation of Related Structures:
3MJ6, 3MJ7 - PubMed Abstract:
Coxsackie and adenovirus receptor (CAR) is the primary cellular receptor for group B coxsackieviruses and most adenovirus serotypes and plays a crucial role in adenoviral gene therapy. Recent discovery of the interaction between junctional adhesion molecule-like protein (JAML) and CAR uncovered important functional roles in immunity, inflammation, and tissue homeostasis. Crystal structures of JAML ectodomain (2.2 angstroms) and its complex with CAR (2.8 angstroms) reveal an unusual immunoglobulin-domain assembly for JAML and a charged interface that confers high specificity. Biochemical and mutagenesis studies illustrate how CAR-mediated clustering of JAML recruits phosphoinositide 3-kinase (P13K) to a JAML intracellular sequence motif as delineated for the alphabeta T cell costimulatory receptor CD28. Thus, CAR and JAML are cell signaling receptors of the immune system with implications for asthma, cancer, and chronic nonhealing wounds.
- Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: