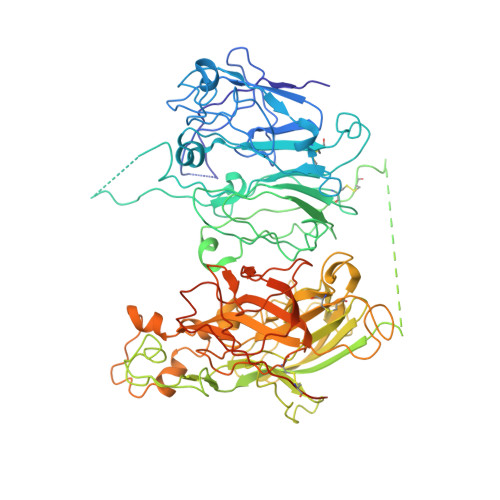
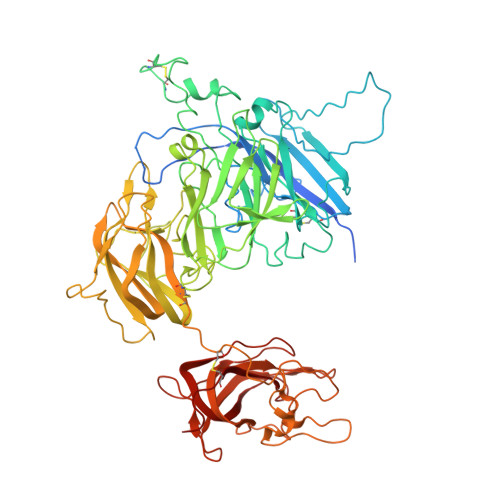
Domain organization of membrane-bound factor VIII.
Stoilova-McPhie, S., Lynch, G.C., Ludtke, S., Pettitt, B.M.(2013) Biopolymers 99: 448-459
- PubMed: 23616213
- DOI: https://doi.org/10.1002/bip.22199
- Primary Citation of Related Structures:
3J2Q, 3J2S - PubMed Abstract:
Factor VIII (FVIII) is the blood coagulation protein which when defective or deficient causes for hemophilia A, a severe hereditary bleeding disorder. Activated FVIII (FVIIIa) is the cofactor to the serine protease factor IXa (FIXa) within the membrane-bound Tenase complex, responsible for amplifying its proteolytic activity more than 100,000 times, necessary for normal clot formation. FVIII is composed of two noncovalently linked peptide chains: a light chain (LC) holding the membrane interaction sites and a heavy chain (HC) holding the main FIXa interaction sites. The interplay between the light and heavy chains (HCs) in the membrane-bound state is critical for the biological efficiency of FVIII. Here, we present our cryo-electron microscopy (EM) and structure analysis studies of human FVIII-LC, when helically assembled onto negatively charged single lipid bilayer nanotubes. The resolved FVIII-LC membrane-bound structure supports aspects of our previously proposed FVIII structure from membrane-bound two-dimensional (2D) crystals, such as only the C2 domain interacts directly with the membrane. The LC is oriented differently in the FVIII membrane-bound helical and 2D crystal structures based on EM data, and the existing X-ray structures. This flexibility of the FVIII-LC domain organization in different states is discussed in the light of the FVIIIa-FIXa complex assembly and function.
- Department of Neuroscience and Cell Biology, University of Texas Medical Branch, Galveston, TX, USA. svmcphie@utmb.edu
Organizational Affiliation: