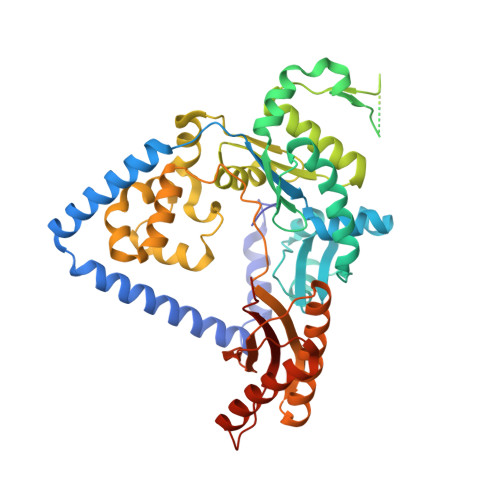
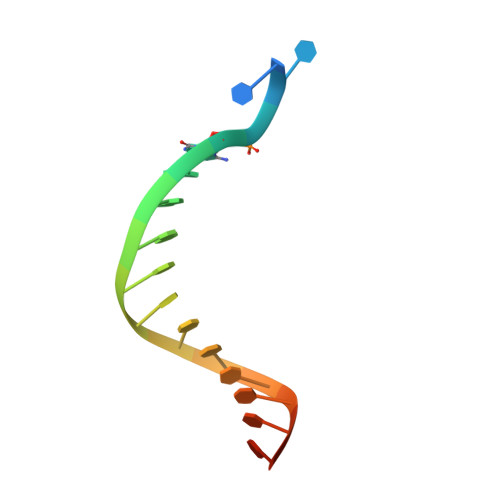
Structure of human DNA polymerase kappa inserting dATP opposite an 8-oxoG DNA lesion
Vasquez-Del Carpio, R., Silverstein, T.D., Lone, S., Swan, M.K., Choudhury, J.R., Johnson, R.E., Prakash, S., Prakash, L., Aggarwal, A.K.(2009) PLoS One 4: e5766-e5766
- PubMed: 19492058
- DOI: https://doi.org/10.1371/journal.pone.0005766
- Primary Citation of Related Structures:
3IN5 - PubMed Abstract:
Oxygen-free radicals formed during normal aerobic cellular metabolism attack bases in DNA and 7,8-dihydro-8-oxoguanine (8-oxoG) is one of the major lesions formed. It is amongst the most mutagenic lesions in cells because of its dual coding potential, wherein 8-oxoG(syn) can pair with an A in addition to normal base pairing of 8-oxoG(anti) with a C. Human DNA polymerase kappa (Polkappa) is a member of the newly discovered Y-family of DNA polymerases that possess the ability to replicate through DNA lesions. To understand the basis of Polkappa's preference for insertion of an A opposite 8-oxoG lesion, we have solved the structure of Polkappa in ternary complex with a template-primer presenting 8-oxoG in the active site and with dATP as the incoming nucleotide. We show that the Polkappa active site is well-adapted to accommodate 8-oxoG in the syn conformation. That is, the polymerase and the bound template-primer are almost identical in their conformations to that in the ternary complex with undamaged DNA. There is no steric hindrance to accommodating 8-oxoG in the syn conformation for Hoogsteen base-paring with incoming dATP. The structure we present here is the first for a eukaryotic translesion synthesis (TLS) DNA polymerase with an 8-oxoG:A base pair in the active site. The structure shows why Polkappa is more efficient at inserting an A opposite the 8-oxoG lesion than a C. The structure also provides a basis for why Polkappa is more efficient at inserting an A opposite the lesion than other Y-family DNA polymerases.
- Department of Structural & Chemical Biology, Mount Sinai School of Medicine, New York, NY, USA.
Organizational Affiliation: