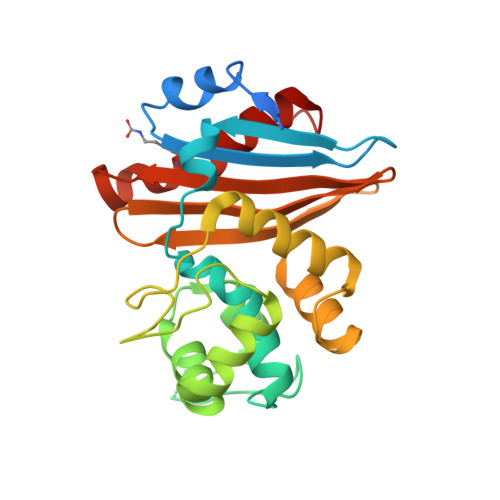
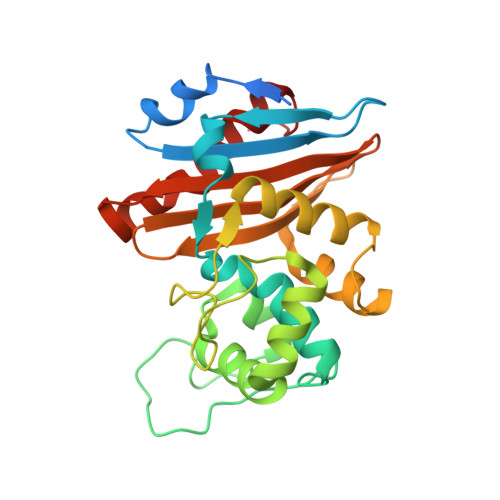
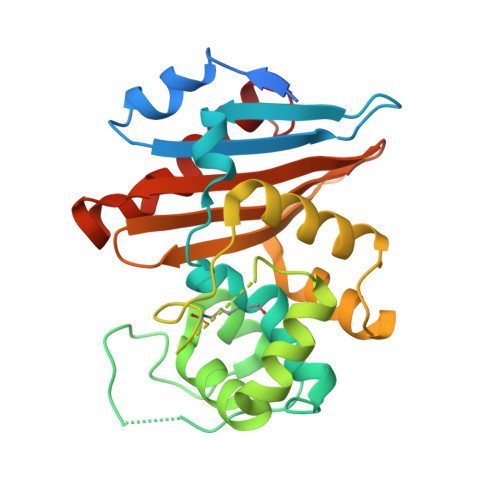
Crystal structure of the narrow-spectrum OXA-46 class D beta-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates
Docquier, J.D., Benvenuti, M., Calderone, V., Giuliani, F., Kapetis, D., De Luca, F., Rossolini, G.M., Mangani, S.(2010) Antimicrob Agents Chemother 54: 2167-2174
- PubMed: 20145076
- DOI: https://doi.org/10.1128/AAC.01517-09
- Primary Citation of Related Structures:
3IF6 - PubMed Abstract:
Class D beta-lactamases represent a heterogeneous group of active-site serine beta-lactamases that show an extraordinary panel of functional features and substrate profiles, thus representing relevant models for biochemical and structural studies. OXA-46 is a narrow-spectrum enzyme belonging to the OXA-2 subgroup which was found in a Pseudomonas aeruginosa clinical isolate from northern Italy. In this work, we obtained the three-dimensional structure of OXA-46, which shows the overall fold of active serine beta-lactamases and a dimeric quaternary structure. Significant differences with currently available structures of class D beta-lactamases were found in the loops located close to the active site, which differ in length and conformation. Interestingly, the three subunits present in the asymmetric unit showed some structural heterogeneity, only one of which presented a carbamylated lysine recognized as an important functional feature of class D enzymes. The carbamylation state of residue Lys75 appeared to be associated with different shapes and dimensions of the active site. Moreover, a tartrate molecule from the crystallization buffer was found in the active site of the noncarbamylated subunits, which interacts with catalytically relevant residues. The OXA-46 crystal asymmetric units thus interestingly present the structures of the free carbamylated active site and of the ligand-bound uncarbamylated active site, offering the structural basis for investigating the potential of new scaffolds of beta-lactamase inhibitors.
- Dipartimento di Biologia Molecolare, Università di Siena, Siena, Italy.
Organizational Affiliation: