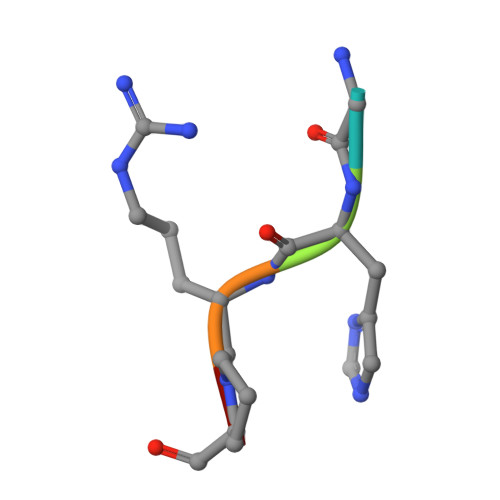
Fibrinogen variant BbetaD432A has normal polymerization but does not bind knob "B".
Bowley, S.R., Lord, S.T.(2009) Blood 113: 4425-4430
- PubMed: 19075185
- DOI: https://doi.org/10.1182/blood-2008-09-178178
- Primary Citation of Related Structures:
3E1I - PubMed Abstract:
Fibrinogen residue Bbeta432Asp is part of hole "b" that interacts with knob "B," whose sequence starts with Gly-His-Arg-Pro-amide (GHRP). Because previous studies showed BbetaD432A has normal polymerization, we hypothesized that Bbeta432Asp is not critical for knob "B" binding and that new knob-hole interactions would compensate for the loss of this Asp residue. To test this hypothesis, we solved the crystal structure of fragment D from BbetaD432A. Surprisingly, the structure (rfD-BbetaD432A+GH) showed the peptide GHRP was not bound to hole "b." We then re-evaluated the polymerization of this variant by examining clot turbidity, clot structure, and the rate of FXIIIa cross-linking. The turbidity and the rate of gamma-gamma dimer formation for BbetaD432A were indistinguishable compared with normal fibrinogen. Scanning electron microscopy showed no significant differences between the clots of BbetaD432A and normal, but the thrombin-derived clots had thicker fibers than clots obtained from batroxobin, suggesting that cleavage of FpB is more important than "B:b" interactions. We conclude that hole "b" and "B:b" knob-hole binding per se have no influence on fibrin polymerization.
- Department of Chemistry, University of North Carolina, Chapel Hill, NC 27599-7525, USA.
Organizational Affiliation: