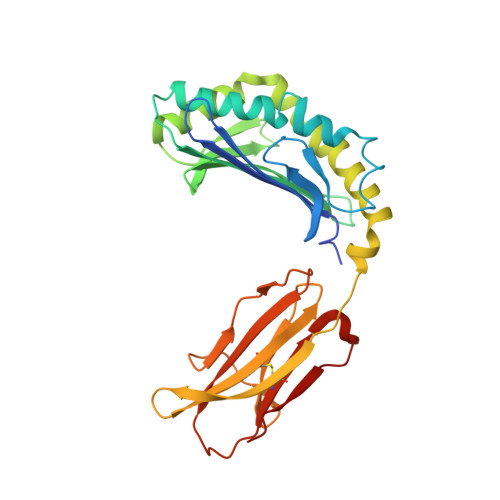
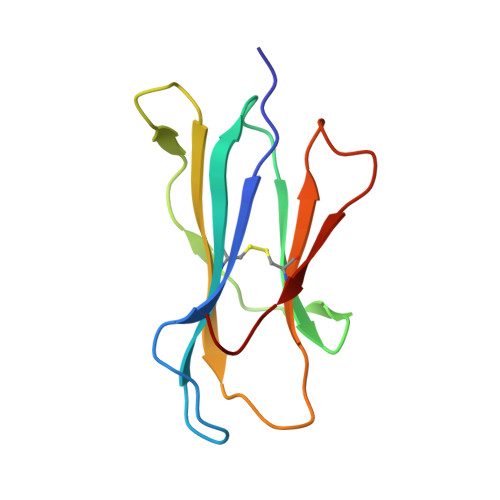
The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1
Zajonc, D.M., Striegl, H., Dascher, C.C., Wilson, I.A.(2008) Proc Natl Acad Sci U S A 105: 17925-17930
- PubMed: 19004781
- DOI: https://doi.org/10.1073/pnas.0809814105
- Primary Citation of Related Structures:
3DBX - PubMed Abstract:
The molecular details of glycolipid presentation by CD1 antigen-presenting molecules are well studied in mammalian systems. However, little is known about how these non-classical MHC class I (MHCI) molecules diverged from the MHC locus to create a more complex, hydrophobic binding groove that binds lipids rather than peptides. To address this fundamental question, we have determined the crystal structure of an avian CD1 (chCD1-2) that shares common ancestry with mammalian CD1 from approximately 310 million years ago. The chCD1-2 antigen-binding site consists of a compact, narrow, central hydrophobic groove or pore rather than the more open, 2-pocket architecture observed in mammalian CD1s. Potential antigens then would be restricted in size to single-chain lipids or glycolipids. An endogenous ligand, possibly palmitic acid, serves to illuminate the mode and mechanism of ligand interaction with chCD1-2. The palmitate alkyl chain is inserted into the relatively shallow hydrophobic pore; its carboxyl group emerges at the receptor surface and is stabilized by electrostatic and hydrogen bond interactions with an arginine residue that is conserved in all known CD1 proteins. In addition, other novel features, such as an A' loop that interrupts and segments the normally long, continuous alpha1 helix, are unique to chCD1-2 and contribute to the unusually narrow binding groove, thereby limiting its size. Because birds and mammals share a common ancestor, but the rate of evolution is slower in birds than in mammals, the chCD1-2-binding groove probably represents a more primordial CD1-binding groove.
- Division of Cell Biology, La Jolla Institute for Allergy and Immunology, 9420 Athena Circle, La Jolla, CA 92037, USA. dzajonc@liai.org
Organizational Affiliation: