Light chain somatic mutations change thermodynamics of binding and water coordination in the HyHEL-10 family of antibodies.
Acchione, M., Lipschultz, C.A., DeSantis, M.E., Shanmuganathan, A., Li, M., Wlodawer, A., Tarasov, S., Smith-Gill, S.J.(2009) Mol Immunol 47: 457-464
- PubMed: 19781789
- DOI: https://doi.org/10.1016/j.molimm.2009.08.018
- Primary Citation of Related Structures:
3D9A - PubMed Abstract:
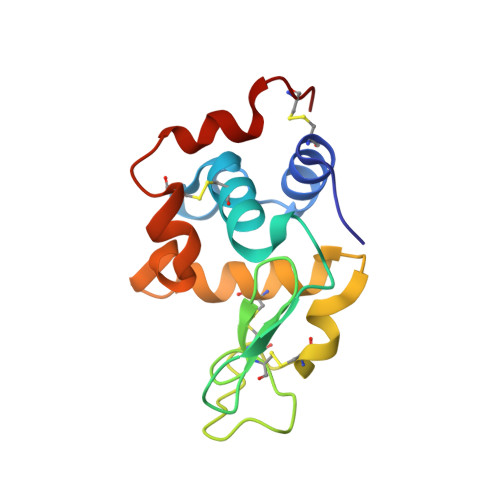
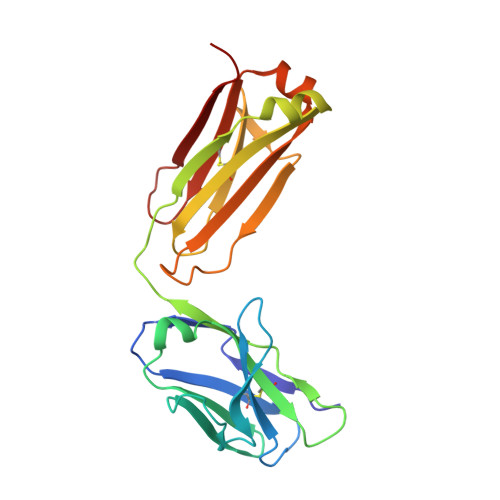
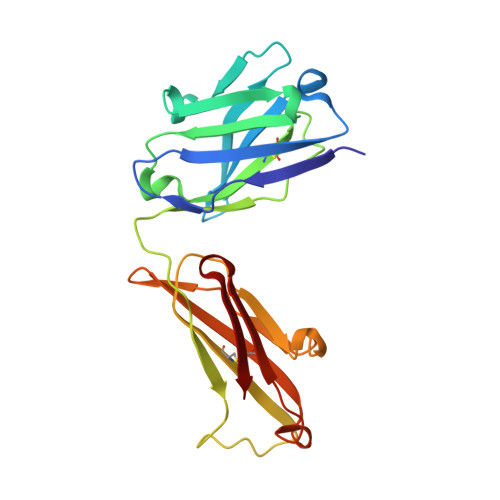
Thermodynamic and structural studies addressed the increased affinity due to L-chain somatic mutations in the HyHEL-10 family of affinity matured IgG antibodies, using ITC, SPR with van't Hoff analysis, and X-ray crystallography. When compared to the parental antibody H26L26, the H26L10 and H26L8 chimeras binding to lysozyme showed an increase in favorable DeltaG(o) of -1.2+/-0.1 kcal mol(-1) and -1.3+/-0.1 kcal mol(-1), respectively. Increase in affinity of the H26L10 chimera was due to a net increase in favorable enthalpy change with little difference in change in entropy compared to H26L26. The H26L8 chimera exhibited the greatest increase in favorable enthalpy but also showed an increase in unfavorable entropy change, with the result being that the affinities of both chimeras were essentially equivalent. Site-directed L-chain mutants identified the shared somatic mutation S30G as the dominant contributor to increasing affinity to lysozyme. This mutation was not influenced by H-chain somatic mutations. Residue 30L is at the periphery of the binding interface and S30G effects an increase in hydrophobicity and decrease in H-bonding ability and size, but does not make any new energetically important antigen contacts. A new 1.2-A structure of the H10L10-HEL complex showed changes in the pattern of both inter- and intra-molecular water bridging with no other significant structural alterations near the binding interface compared to the H26L26-HEL complex. These results highlight the necessity for investigating both the structure and the thermodynamics associated with introduced mutations, in order to better assess and understand their impact on binding. Furthermore, it provides an important example of how backbone flexibility and water-bridging may favorably influence the thermodynamics of an antibody-antigen interaction.
- Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD, USA.
Organizational Affiliation: