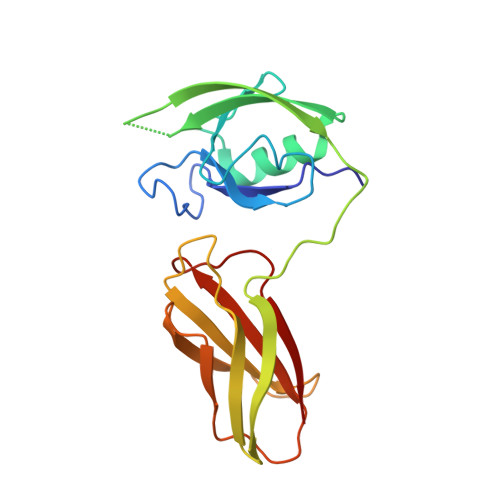
Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution.
Huang, J., Makabe, K., Biancalana, M., Koide, A., Koide, S.(2009) J Mol Biology 392: 1221-1231
- PubMed: 19646997
- DOI: https://doi.org/10.1016/j.jmb.2009.07.067
- Primary Citation of Related Structures:
3CH8 - PubMed Abstract:
We have established a new protein-engineering strategy termed "directed domain-interface evolution" that generates a binding site by linking two protein domains and then optimizing the interface between them. Using this strategy, we have generated synthetic two-domain "affinity clamps" using PDZ and fibronectin type III (FN3) domains as the building blocks. While these affinity clamps all had significantly higher affinity toward a target peptide than the underlying PDZ domain, two distinct types of affinity clamps were found in terms of target specificity. One type conserved the specificity of the parent PDZ domain, and the other increased the specificity dramatically. Here, we characterized their specificity profiles using peptide phage-display libraries and scanning mutagenesis, which suggested a significantly enlarged recognition site of the high-specificity affinity clamps. The crystal structure of a high-specificity affinity clamp showed extensive contacts with a portion of the peptide ligand that is not recognized by the parent PDZ domain, thus rationalizing the improvement of the specificity of the affinity clamp. A comparison with another affinity clamp structure showed that, although both had extensive contacts between PDZ and FN3 domains, they exhibited a large offset in the relative position of the two domains. Our results indicate that linked domains could rapidly fuse and evolve as a single functional module, and that the inherent plasticity of domain interfaces allows for the generation of diverse active-site topography. These attributes of directed domain-interface evolution provide facile means to generate synthetic proteins with a broad range of functions.
- Department of Biochemistry and Molecular Biology, The University of Chicago, 929 East 57th Street, Chicago, IL 60637, USA.
Organizational Affiliation: