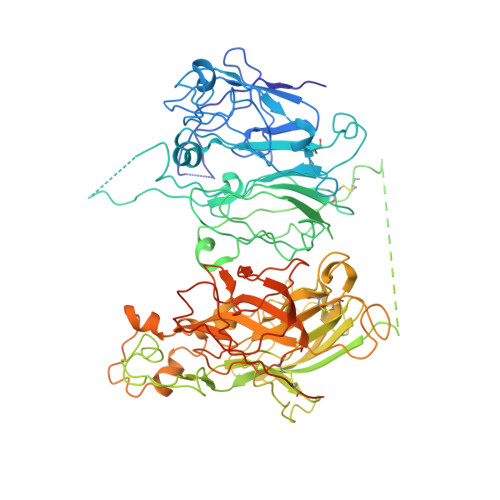
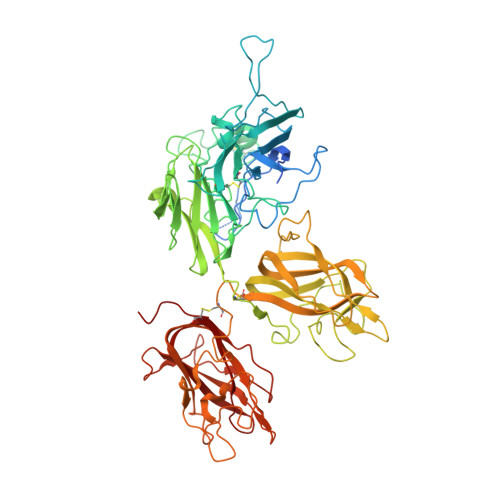
Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex.
Ngo, J.C., Huang, M., Roth, D.A., Furie, B.C., Furie, B.(2008) Structure 16: 597-606
- PubMed: 18400180
- DOI: https://doi.org/10.1016/j.str.2008.03.001
- Primary Citation of Related Structures:
3CDZ - PubMed Abstract:
Factor VIII is a procofactor that plays a critical role in blood coagulation, and is missing or defective in hemophilia A. We determined the X-ray crystal structure of B domain-deleted human factor VIII. This protein is composed of five globular domains and contains one Ca(2+) and two Cu(2+) ions. The three homologous A domains form a triangular heterotrimer where the A1 and A3 domains serve as the base and interact with the C2 and C1 domains, respectively. The structurally homologous C1 and C2 domains reveal membrane binding features. Based on biochemical studies, a model of the factor IXa-factor VIIIa complex was constructed by in silico docking. Factor IXa wraps across the side of factor VIII, and an extended interface spans the factor VIII heavy and light chains. This model provides insight into the activation of factor VIII and the interaction of factor VIIIa with factor IXa on the membrane surface.
- Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Organizational Affiliation: