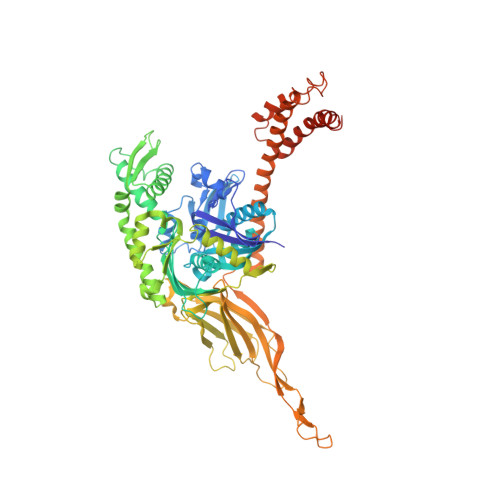
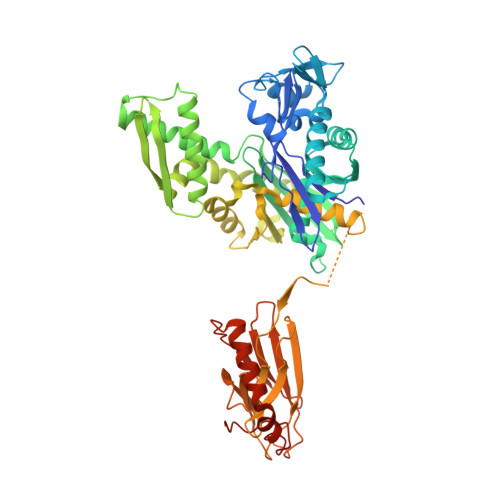
Structure of the Hsp110:Hsc70 nucleotide exchange machine
Schuermann, J.P., Jiang, J., Cuellar, J., Llorca, O., Wang, L., Gimenez, L.E., Jin, S., Taylor, A.B., Demeler, B., Morano, K.A., Hart, P.J., Valpuesta, J.M., Lafer, E.M., Sousa, R.(2008) Mol Cell 31: 232-243
- PubMed: 18550409
- DOI: https://doi.org/10.1016/j.molcel.2008.05.006
- Primary Citation of Related Structures:
3C7N - PubMed Abstract:
Hsp70s mediate protein folding, translocation, and macromolecular complex remodeling reactions. Their activities are regulated by proteins that exchange ADP for ATP from the nucleotide-binding domain (NBD) of the Hsp70. These nucleotide exchange factors (NEFs) include the Hsp110s, which are themselves members of the Hsp70 family. We report the structure of an Hsp110:Hsc70 nucleotide exchange complex. The complex is characterized by extensive protein:protein interactions and symmetric bridging interactions between the nucleotides bound in each partner protein's NBD. An electropositive pore allows nucleotides to enter and exit the complex. The role of nucleotides in complex formation and dissociation, and the effects of the protein:protein interactions on nucleotide exchange, can be understood in terms of the coupled effects of the nucleotides and protein:protein interactions on the open-closed isomerization of the NBDs. The symmetrical interactions in the complex may model other Hsp70 family heterodimers in which two Hsp70s reciprocally act as NEFs.
- Department of Biochemistry, University of Texas Health Science Center, San Antonio, TX 78229-3900, USA.
Organizational Affiliation: