Reaction mechanism of glutamate carboxypeptidase II revealed by mutagenesis, X-ray crystallography, and computational methods.
Klusak, V., Barinka, C., Plechanovova, A., Mlcochova, P., Konvalinka, J., Rulisek, L., Lubkowski, J.(2009) Biochemistry 48: 4126-4138
- PubMed: 19301871
- DOI: https://doi.org/10.1021/bi900220s
- Primary Citation of Related Structures:
3BXM - PubMed Abstract:
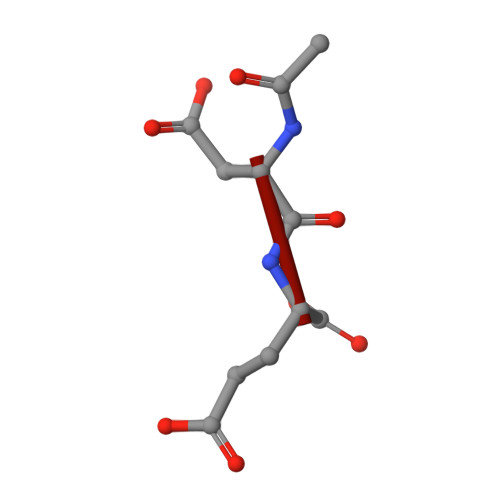
Glutamate carboxypeptidase II (GCPII, EC 3.4.17.21) is a zinc-dependent exopeptidase and an important therapeutic target for neurodegeneration and prostate cancer. The hydrolysis of N-acetyl-l-aspartyl-l-glutamate (N-Ac-Asp-Glu), the natural dipeptidic substrate of the GCPII, is intimately involved in cellular signaling within the mammalian nervous system, but the exact mechanism of this reaction has not yet been determined. To investigate peptide hydrolysis by GCPII in detail, we constructed a mutant of human GCPII [GCPII(E424A)], in which Glu424, a putative proton shuttle residue, is substituted with alanine. Kinetic analysis of GCPII(E424A) using N-Ac-Asp-Glu as substrate revealed a complete loss of catalytic activity, suggesting the direct involvement of Glu424 in peptide hydrolysis. Additionally, we determined the crystal structure of GCPII(E424A) in complex with N-Ac-Asp-Glu at 1.70 A resolution. The presence of the intact substrate in the GCPII(E424A) binding cavity substantiates our kinetic data and allows a detailed analysis of GCPII/N-Ac-Asp-Glu interactions. The experimental data are complemented by the combined quantum mechanics/molecular mechanics calculations (QM/MM) which enabled us to characterize the transition states, including the associated reaction barriers, and provided detailed information concerning the GCPII reaction mechanism. The best estimate of the reaction barrier was calculated to be DeltaG(++) approximately 22(+/-5) kcal x mol(-1), which is in a good agreement with the experimentally observed reaction rate constant (k(cat) approximately 1 s(-1)). Combined together, our results provide a detailed and consistent picture of the reaction mechanism of this highly interesting enzyme at the atomic level.
- Institute of Organic Chemistry and Biochemistry, Gilead Sciences Research Center and IOCB, Academy of Sciences of the Czech Republic, Flemingovo nam. 2, 166 10 Praha 6, Czech Republic.
Organizational Affiliation: