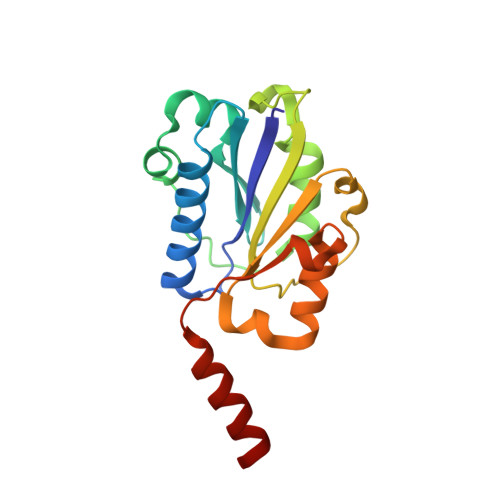
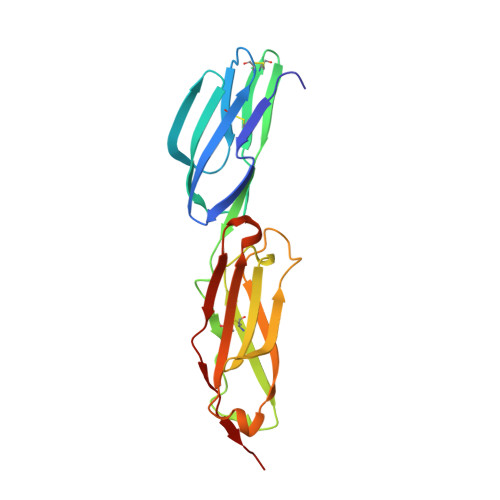
An unusual allosteric mobility of the C-terminal helix of a high-affinity alpha L integrin I domain variant bound to ICAM-5
Zhang, H., Casasnovas, J.M., Jin, M., Liu, J.H., Gahmberg, C.G., Springer, T.A., Wang, J.-h.(2008) Mol Cell 31: 432-437
- PubMed: 18691975
- DOI: https://doi.org/10.1016/j.molcel.2008.06.022
- Primary Citation of Related Structures:
3BN3 - PubMed Abstract:
Integrins are cell surface receptors that transduce signals bidirectionally across the plasma membrane. The key event of integrin signaling is the allosteric regulation between its ligand-binding site and the C-terminal helix (alpha7) of integrin's inserted (I) domain. A significant axial movement of the alpha7 helix is associated with the open, active conformation of integrins. We describe the crystal structure of an engineered high-affinity I domain from the integrin alpha(L)beta(2) (LFA-1) alpha subunit in complex with the N-terminal two domains of ICAM-5, an adhesion molecule expressed in telencephalic neurons. The finding that the alpha7 helix swings out and inserts into a neighboring I domain in an upside-down orientation in the crystals implies an intrinsically unusual mobility of this helix. This remarkable feature allows the alpha7 helix to trigger integrin's large-scale conformational changes with little energy penalty. It serves as a mechanistic example of how a weakly bound adhesion molecule works in signaling.
- Department of Medical Oncology and Department of Cancer Biology, Dana-Farber Cancer Institute, 44 Binney Street, Boston, MA 02115, USA.
Organizational Affiliation: