A direct crystallographic demonstration that Type II restriction endonuclease PspGI flips nucleotides
Szczepanowski, R.H., Carpenter, M., Czapinska, H., Tamulaitis, G., Siksnys, V., Bhagwat, A., Bochtler, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PspGI restriction endonuclease | C [auth A], D [auth B] | 272 | Pyrococcus sp. GI-H | Mutation(s): 2 | 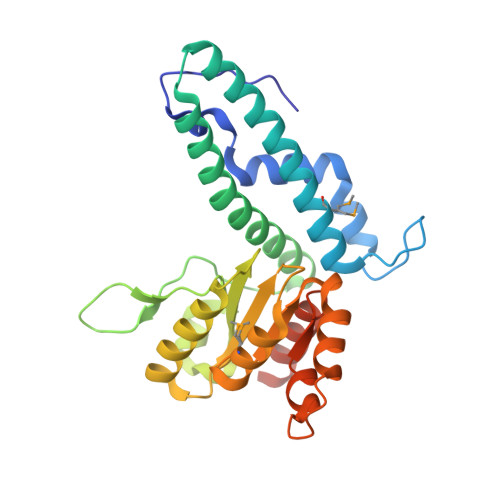 |
UniProt | |||||
Find proteins for O93646 (Pyrococcus sp. GI-H) Explore O93646 Go to UniProtKB: O93646 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O93646 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3') | A [auth C] | 11 | N/A | 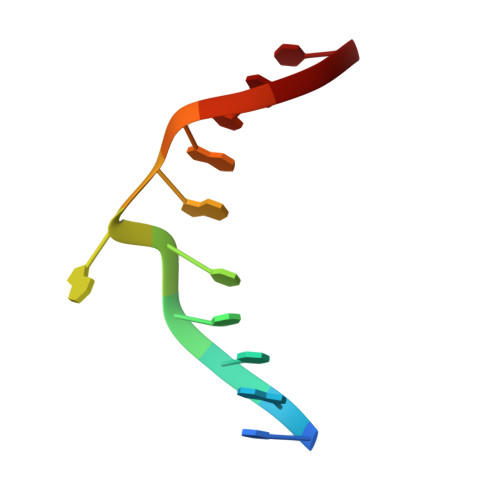 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3') | B [auth D] | 11 | N/A | 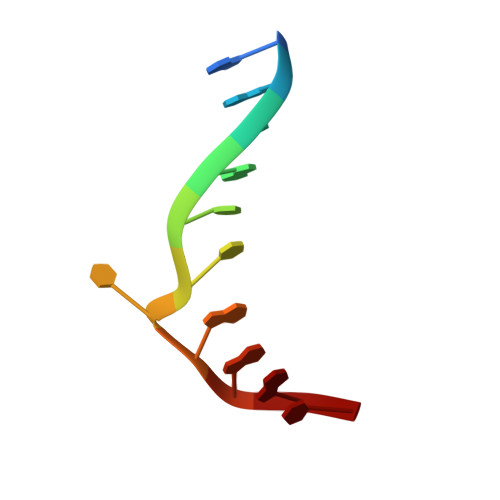 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CIT Query on CIT | E [auth B] | CITRIC ACID C6 H8 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | C [auth A], D [auth B] | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 46.884 | α = 90 |
| b = 96.06 | β = 90 |
| c = 127.069 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SHELXD | phasing |
| SHELXE | model building |
| DM | model building |
| ARP/wARP | model building |
| REFMAC | refinement |
| MAR345 | data collection |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| DM | phasing |