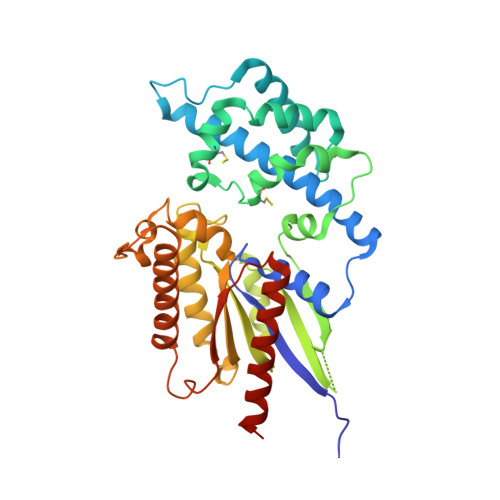
The Crystal Structure of a Self-Activating G Protein Alpha Subunit Reveals its Distinct Mechanism of Signal Initiation
Jones, J.C., Duffy, J.W., Machius, M., Temple, B.R.S., Dohlman, H.G., Jones, A.M.(2011) Sci Signal 159: RA8
- PubMed: 21304159
- DOI: https://doi.org/10.1126/scisignal.2001446
- Primary Citation of Related Structures:
2XTZ - PubMed Abstract:
In animals, heterotrimeric guanine nucleotide-binding protein (G protein) signaling is initiated by G protein-coupled receptors (GPCRs), which activate G protein α subunits; however, the plant Arabidopsis thaliana lacks canonical GPCRs, and its G protein α subunit (AtGPA1) is self-activating. To investigate how AtGPA1 becomes activated, we determined its crystal structure. AtGPA1 is structurally similar to animal G protein α subunits, but our crystallographic and biophysical studies revealed that it had distinct properties. Notably, the helical domain of AtGPA1 displayed pronounced intrinsic disorder and a tendency to disengage from the Ras domain of the protein. Domain substitution experiments showed that the helical domain of AtGPA1 was necessary for self-activation and sufficient to confer self-activation to an animal G protein α subunit. These findings reveal the structural basis for a mechanism for G protein activation in Arabidopsis that is distinct from the well-established mechanism found in animals.
- Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599, USA.
Organizational Affiliation: