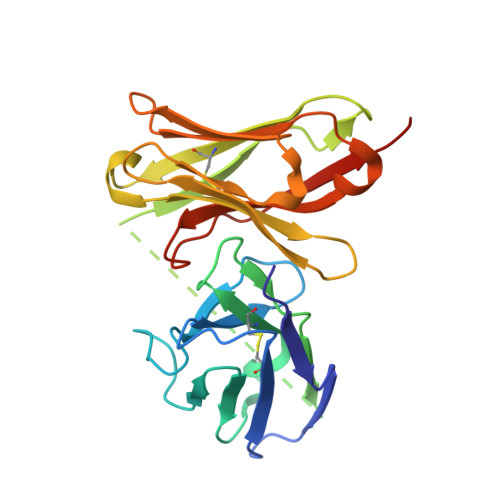
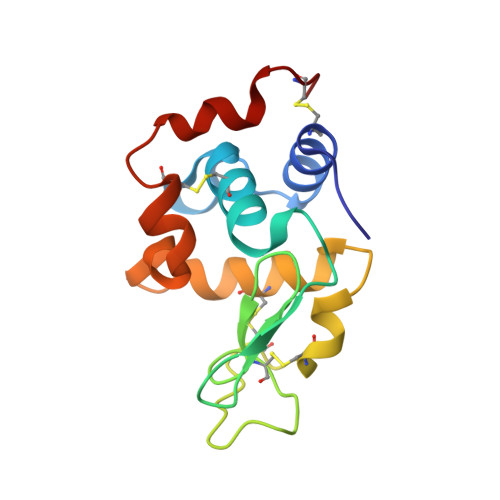
Specific fluorine labeling of the HyHEL10 antibody affects antigen binding and dynamics
Acchione, M., Lee, Y.C., DeSantis, M.E., Lipschultz, C.A., Wlodawer, A., Li, M., Shanmuganathan, A., Walter, R.L., Smith-Gill, S., Barchi, J.J.(2012) Biochemistry 51: 6017-6027
- PubMed: 22769726
- DOI: https://doi.org/10.1021/bi300455t
- Primary Citation of Related Structures:
2ZNW, 2ZNX - PubMed Abstract:
To more fully understand the molecular mechanisms responsible for variations in binding affinity with antibody maturation, we explored the use of site specific fluorine labeling and (19)F nuclear magnetic resonance (NMR). Several single-chain (scFv) antibodies, derived from an affinity-matured series of anti-hen egg white lysozyme (HEL) mouse IgG1, were constructed with either complete or individual replacement of tryptophan residues with 5-fluorotryptophan ((5F)W). An array of biophysical techniques was used to gain insight into the impact of fluorine substitution on the overall protein structure and antigen binding. SPR measurements indicated that (5F)W incorporation lowered binding affinity for the HEL antigen. The degree of analogue impact was residue-dependent, and the greatest decrease in affinity was observed when (5F)W was substituted for residues near the binding interface. In contrast, corresponding crystal structures in complex with HEL were essentially indistinguishable from the unsubstituted antibody. (19)F NMR analysis showed severe overlap of signals in the free fluorinated protein that was resolved upon binding to antigen, suggesting very distinct chemical environments for each (5F)W in the complex. Preliminary relaxation analysis suggested the presence of chemical exchange in the antibody-antigen complex that could not be observed by X-ray crystallography. These data demonstrate that fluorine NMR can be an extremely useful tool for discerning structural changes in scFv antibody-antigen complexes with altered function that may not be discernible by other biophysical techniques.
- Structural Biophysics Laboratory, Center for Cancer Research, Frederick National Laboratory for Cancer Research, Frederick, MD 21702, USA.
Organizational Affiliation: