Conformational Changes in Ige Contribute to its Uniquely Slow Dissociation Rate from Receptor Fceri
Holdom, M.D., Davies, A.M., Nettleship, J.E., Bagby, S.C., Dhaliwal, B., Girardi, E., Hunt, J., Gould, H.J., Beavil, A.J., Mcdonnell, J.M., Owens, R.J., Sutton, B.J.(2011) Nat Struct Mol Biol 18: 571
- PubMed: 21516097
- DOI: https://doi.org/10.1038/nsmb.2044
- Primary Citation of Related Structures:
2WQR, 2Y7Q - PubMed Abstract:
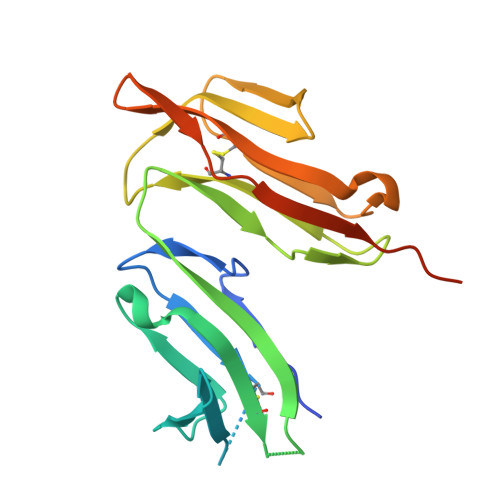
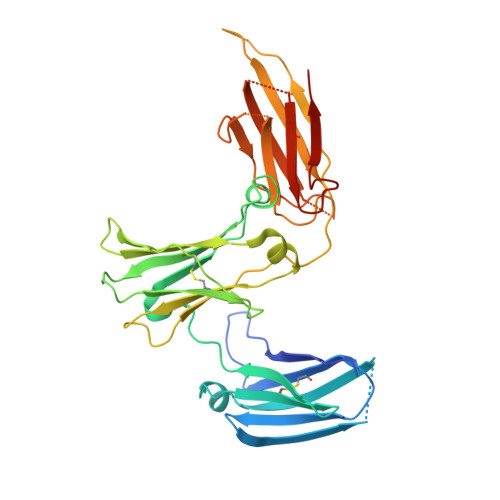
Among antibody classes, IgE has a uniquely slow dissociation rate from, and high affinity for, its cell surface receptor FcɛRI. We show the structural basis for these key determinants of the ability of IgE to mediate allergic hypersensitivity through the 3.4-Å-resolution crystal structure of human IgE-Fc (consisting of the Cɛ2, Cɛ3 and Cɛ4 domains) bound to the extracellular domains of the FcɛRI α chain. Comparison with the structure of free IgE-Fc (reported here at a resolution of 1.9 Å) shows that the antibody, which has a compact, bent structure before receptor engagement, becomes even more acutely bent in the complex. Thermodynamic analysis indicates that the interaction is entropically driven, which explains how the noncontacting Cɛ2 domains, in place of the flexible hinge region of IgG antibodies, contribute together with the conformational changes to the unique binding properties of IgE.
- King's College London, Randall Division of Cell and Molecular Biophysics, London, UK.
Organizational Affiliation: