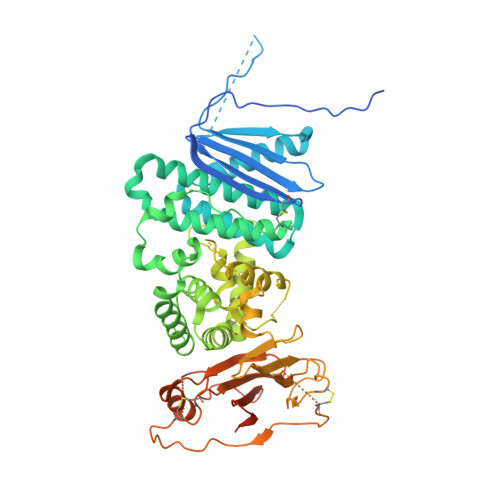
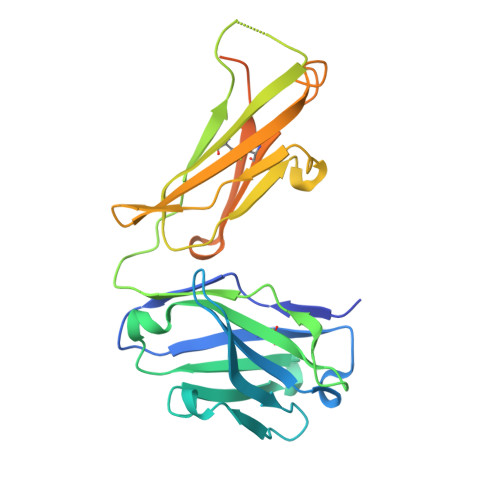
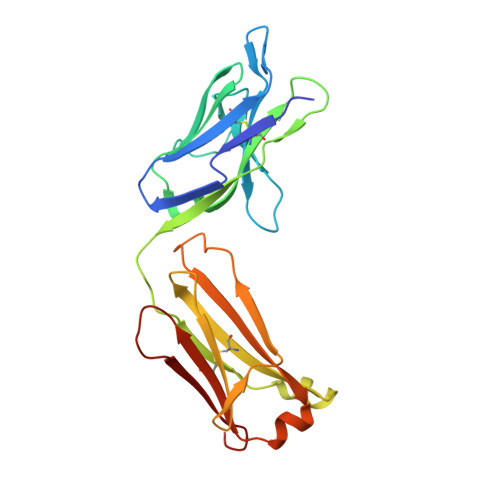
Structure of a Core Fragment of Glycoprotein H from Pseudorabies Virus in Complex with Antibody.
Backovic, M., Dubois, R.M., Cockburn, J.J., Sharff, A.J., Vaney, M.C., Granzow, H., Klupp, B.G., Bricogne, G., Mettenleiter, T.C., Rey, F.A.(2010) Proc Natl Acad Sci U S A 107: 22635
- PubMed: 21149698
- DOI: https://doi.org/10.1073/pnas.1011507107
- Primary Citation of Related Structures:
2XQY - PubMed Abstract:
Compared with many well-studied enveloped viruses, herpesviruses use a more sophisticated molecular machinery to induce fusion of viral and cellular membranes during cell invasion. This essential function is carried out by glycoprotein B (gB), a class III viral fusion protein, together with the heterodimer of glycoproteins H and L (gH/gL). In pseudorabies virus (PrV), a porcine herpesvirus, it was shown that gH/gL can be substituted by a chimeric fusion protein gDgH, containing the receptor binding domain (RBD) of glycoprotein D fused to a truncated version of gH lacking its N-terminal domain. We report here the 2.1-Å resolution structure of the core fragment of gH present in this chimera, bound to the Fab fragment of a PrV gH-specific monoclonal antibody. The structure strongly complements the information derived from the recently reported structure of gH/gL from herpes simplex virus type 2 (HSV-2). Together with the structure of Epstein-Barr virus (EBV) gH/gL reported in parallel, it provides insight into potentially functional conserved structural features. One feature is the presence of a syntaxin motif, and the other is an extended "flap" masking a conserved hydrophobic patch in the C-terminal domain, which is closest to the viral membrane. The negative electrostatic surface potential of this domain suggests repulsive interactions with the lipid heads. The structure indicates the possible unmasking of an extended hydrophobic patch by movement of the flap during a receptor-triggered conformational change of gH, exposing a hydrophobic surface to interact with the viral membrane during the fusion process.
- Institut Pasteur, Département de Virologie, Unité de Virologie Structurale, 75724 Paris Cedex 15, France.
Organizational Affiliation: