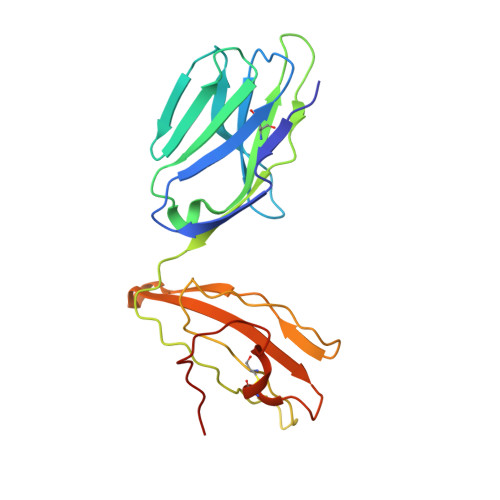
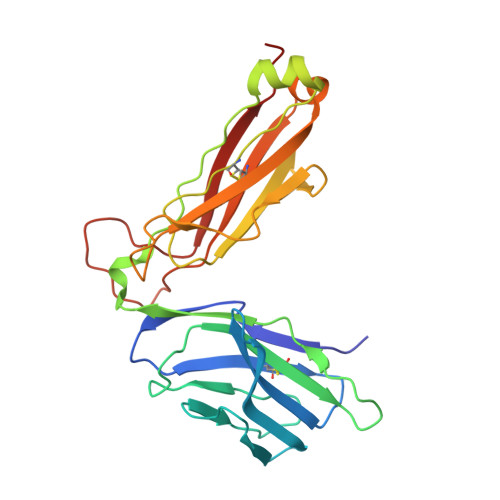
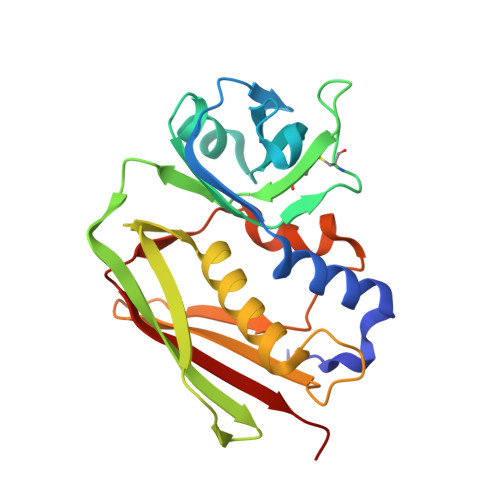
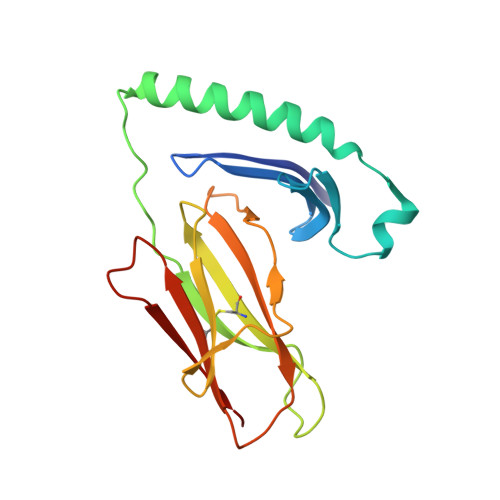
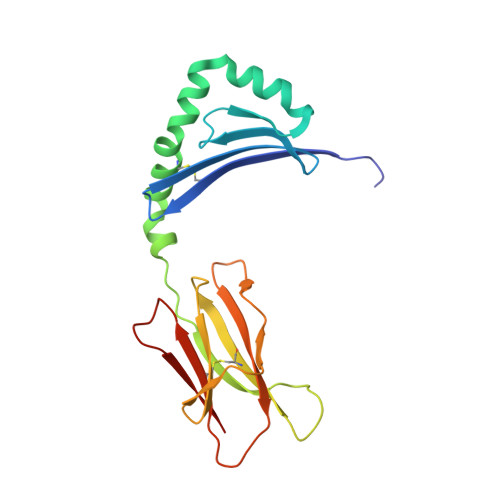
The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Saline, M., Rodstrom, K.E.J., Fischer, G., Orekhov, V.Y., Karlsson, B.G., Lindkvist-Petersson, K.(2010) Nat Commun 1: 119
- PubMed: 21081917
- DOI: https://doi.org/10.1038/ncomms1117
- Primary Citation of Related Structures:
2XN9, 2XNA - PubMed Abstract:
Superantigens (SAgs) are bacterial toxins that interact with immunoreceptors, T cell receptor (TCR) and major histocompatibility complex (MHC) class II, conventionally through the variable β-domain of TCR (TCRVβ). They induce a massive release of cytokines, which can lead to diseases such as food poisoning and toxic shock syndrome. In this study, we report the X-ray structure of the ternary complex between staphylococcal enterotoxin H (SEH) and its human receptors, MHC class II and TCR. The structure demonstrates that SEH predominantly interacts with the variable α-domain of TCR (TCRVα), which is supported by nuclear magnetic resonance (NMR) analyses. Furthermore, there is no contact between MHC and TCR upon complex formation. Structural analyses suggest that the major contact points to TCRVα are conserved among other bacterial SAgs. Consequently, a new dimension of SAg biology emerges, suggesting that in addition to the conventional interactions with the TCRVβ domain, SAgs can also activate T cells through the TCRVα domain.
- 1] Swedish NMR Centre at University of Gothenburg, Box 465, Göteborg S-405 30, Sweden. [2] Department of Chemistry, University of Gothenburg, Box 462, Göteborg S-405 30, Sweden. [3] Swedish NMR Centre at University of Gothenburg, Box 465, Göteborg S-405 30, Sweden. [4].
Organizational Affiliation: