Design, Synthesis, and Evaluation of 2-Methyl- and 2-Amino-N-Aryl-4,5-Dihydrothiazolo[4,5-H]Quinazolin-8-Amines as Ring-Constrained 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Cyclin-Dependent Kinase Inhibitors.
Mcintyre, N.A., Mcinnes, C., Griffiths, G., Barnett, A.L., Kontopidis, G., Slawin, A.M.Z., Jackson, W., Thomas, M., Zheleva, D.I., Wang, S., Blake, D.G., Westwood, N.J., Fischer, P.M.(2010) J Med Chem 53: 2136
- PubMed: 20146435
- DOI: https://doi.org/10.1021/jm901660c
- Primary Citation of Related Structures:
2X1N - PubMed Abstract:
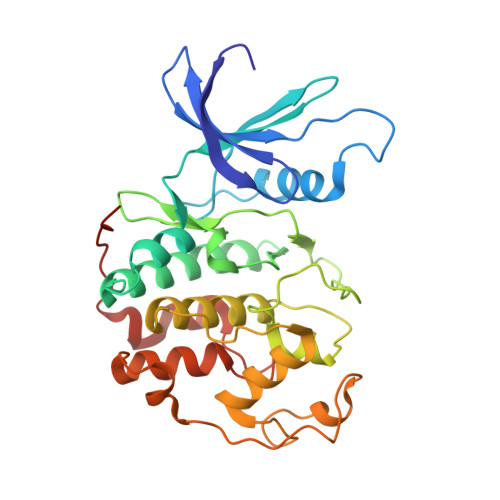
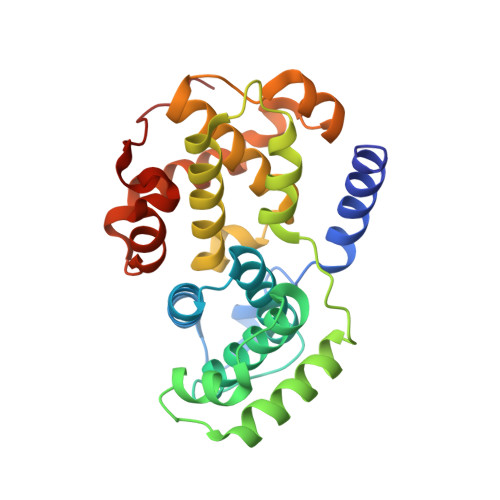
Following the recent discovery and development of 2-anilino-4-(thiazol-5-yl)pyrimidine cyclin dependent kinase (CDK) inhibitors, a program was initiated to evaluate related ring-constrained analogues, specifically, 2-methyl- and 2-amino-N-aryl-4,5-dihydrothiazolo[4,5-h]quinazolin-8-amines for inhibition of CDKs. Here we report the rational design, synthesis, structure-activity relationships (SARs), and cellular mode-of-action profile of these second generation CDK inhibitors. Many of the analogues from this chemical series inhibit CDKs with very low nanomolar K(i) values. The most potent compound reported in this study inhibits CDK2 with an IC(50) of 0.7 nM ([ATP] = 100 microM). Furthermore, an X-ray crystal structure of 2-methyl-N-(3-(nitro)phenyl)-4,5-dihydrothiazolo[4,5-h]quinazolin-8-amine (11g), a representative from the chemical series in complex with cyclin A-CDK2, is reported, confirming the design rationale and expected binding mode within the CDK2 ATP binding pocket.
- School of Chemistry and Biomedical Sciences Research Complex, University of St. Andrews, North Haugh, St Andrews, Fife KY16 9ST, UK.
Organizational Affiliation: