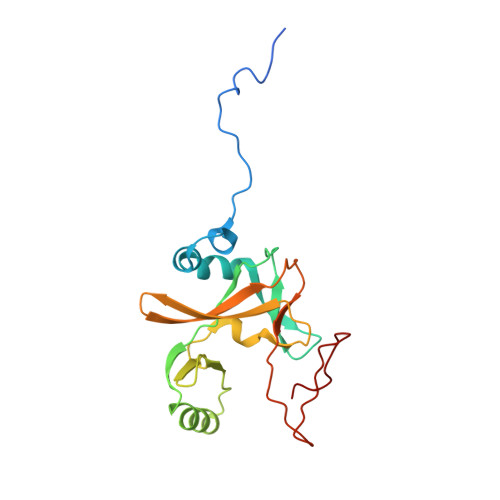
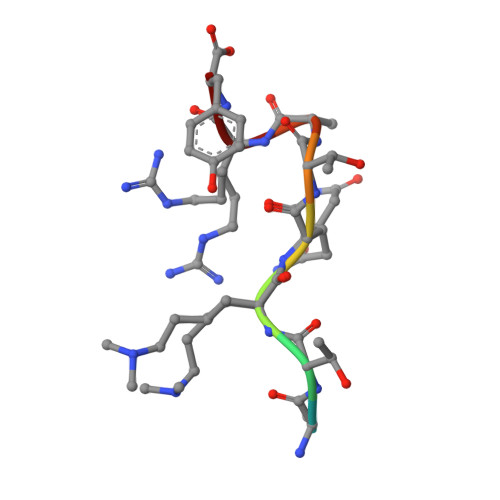
Structural Basis for the Requirement of Additional Factors for Mll1 Set Domain Activity and Recognition of Epigenetic Marks.
Southall, S.M., Wong, P.S., Odho, Z., Roe, S.M., Wilson, J.R.(2009) Mol Cell 33: 181
- PubMed: 19187761
- DOI: https://doi.org/10.1016/j.molcel.2008.12.029
- Primary Citation of Related Structures:
2W5Y, 2W5Z - PubMed Abstract:
The mixed-lineage leukemia protein MLL1 is a transcriptional regulator with an essential role in early development and hematopoiesis. The biological function of MLL1 is mediated by the histone H3K4 methyltransferase activity of the carboxyl-terminal SET domain. We have determined the crystal structure of the MLL1 SET domain in complex with cofactor product AdoHcy and a histone H3 peptide. This structure indicates that, in order to form a well-ordered active site, a highly variable but essential component of the SET domain must be repositioned. To test this idea, we compared the effect of the addition of MLL complex members on methyltransferase activity and show that both RbBP5 and Ash2L but not Wdr5 stimulate activity. Additionally, we have determined the effect of posttranslational modifications on histone H3 residues downstream and upstream from the target lysine and provide a structural explanation for why H3T3 phosphorylation and H3K9 acetylation regulate activity.
- Institute of Cancer Research, Chester Beatty Laboratories, Chelsea, London, UK.
Organizational Affiliation: