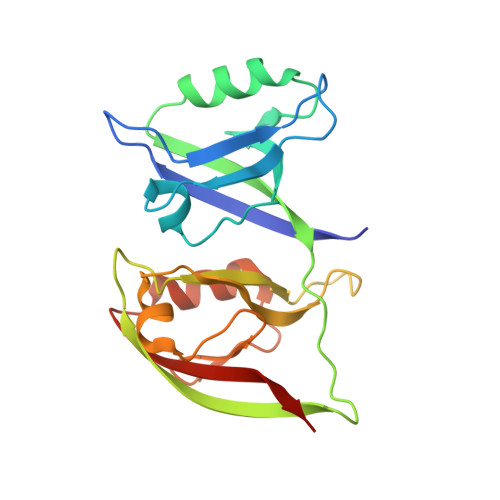
Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1.
Long, J., Wei, Z., Feng, W., Yu, C., Zhao, Y.X., Zhang, M.(2008) J Mol Biology 375: 1457-1468
- PubMed: 18155042
- DOI: https://doi.org/10.1016/j.jmb.2007.11.088
- Primary Citation of Related Structures:
2QT5 - PubMed Abstract:
The scaffold protein GRIP1 (glutamate receptor interacting protein 1) binds to and regulates both the trafficking and membrane organization of a large number of transmembrane proteins. Mutation of GRIP1 in mice displays essentially the same phenotype of the mutations of Fras1 or Frem2, which are the animal models of the human genetic disorder Fraser syndrome. However, the molecular basis governing the interaction between GRIP1 and Fras1/Frem2 is unknown. Here, we show that interaction between Fras1 and GRIP1 requires the first two PDZ domains (PDZ1 and PDZ2) to be connected in tandem, as the folding of PDZ1 strictly depends on the covalent attachment of PDZ2. The crystal structure of GRIP1 PDZ12 in complex with the Fras1 C-terminal peptide reveals that the PDZ12 tandem forms a supramodule in which only the peptide-binding groove of PDZ1 is bound with the Fras1 peptide. The GRIP1 PDZ12/Fras1 peptide complex not only provides a mechanistic explanation of the link between GRIP1 and the Fraser syndrome but may also serve as a foundation for searching for potential mutations in GRIP1 that could lead to the Fraser syndrome.
- Department of Biochemistry, Molecular Neuroscience Center, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong.
Organizational Affiliation: