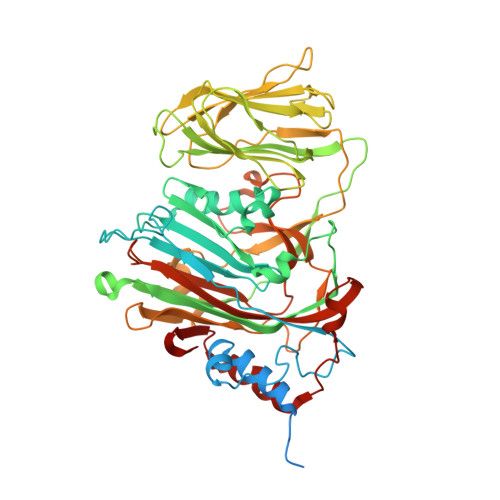
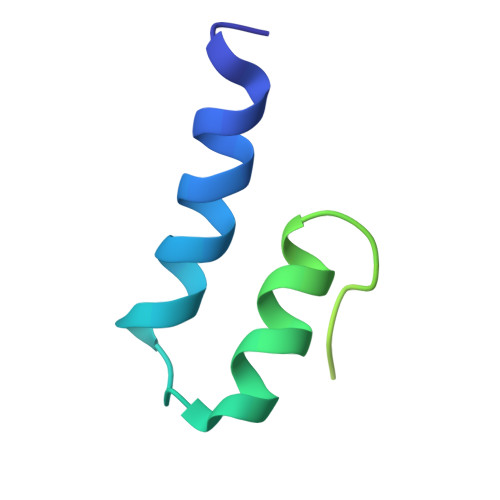
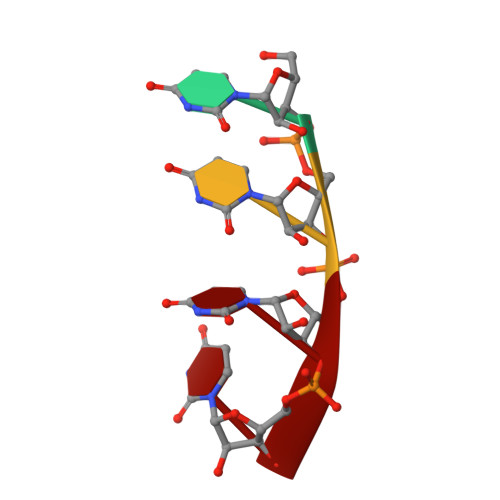
Evolution in action: N and C termini of subunits in related T = 4 viruses exchange roles as molecular switches.
Speir, J.A., Taylor, D.J., Natarajan, P., Pringle, F.M., Ball, L.A., Johnson, J.E.(2010) Structure 18: 700-709
- PubMed: 20541507
- DOI: https://doi.org/10.1016/j.str.2010.03.010
- Primary Citation of Related Structures:
2QQP - PubMed Abstract:
The T = 4 tetravirus and T = 3 nodavirus capsid proteins undergo closely similar autoproteolysis to produce the N-terminal beta and C-terminal, lipophilic gamma polypeptides. The gamma peptides and the N termini of beta also act as molecular switches that determine their quasi equivalent capsid structures. The crystal structure of Providence virus (PrV), only the second of a tetravirus (the first was NomegaV), reveals conserved folds and cleavage sites, but the protein termini have completely different structures and the opposite functions of those in NomegaV. N termini of beta form the molecular switch in PrV, whereas gamma peptides play this role in NomegaV. PrV gamma peptides instead interact with packaged RNA at the particle two-folds by using a repeating sequence pattern found in only four other RNA- or membrane-binding proteins. The disposition of peptide termini in PrV is closely related to those in nodaviruses, suggesting that PrV may be closer to the primordial T = 4 particle than NomegaV.
- Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: