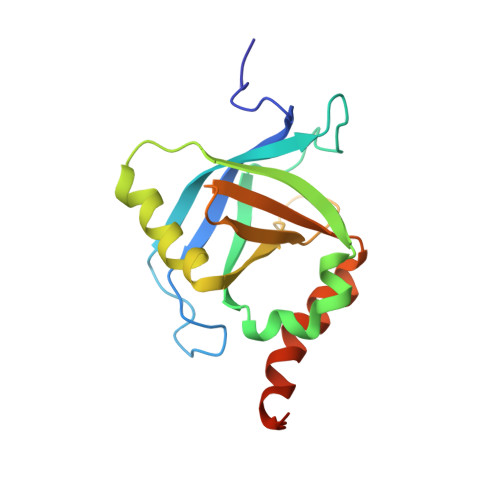
NMR structure of the human Mediator MED25 ACID domain.
Bontems, F., Verger, A., Dewitte, F., Lens, Z., Baert, J.L., Ferreira, E., Launoit, Y., Sizun, C., Guittet, E., Villeret, V., Monte, D.(2011) J Struct Biol 174: 245-251
- PubMed: 20974256
- DOI: https://doi.org/10.1016/j.jsb.2010.10.011
- Primary Citation of Related Structures:
2L23 - PubMed Abstract:
MED25 (ARC92/ACID1) is a 747 residues subunit specific to higher eukaryote Mediator complex, an essential component of the RNA polymerase II general transcriptional machinery. MED25 is a target of the Herpes simplex virus transactivator protein VP16. MED25 interacts with VP16 through a central MED25 PTOV (Prostate tumour overexpressed)/ACID (Activator interacting domain) domain of unknown structure. As a first step towards understanding the mechanism of recruitment of transactivation domains by MED25, we report here the NMR structure of the MED25 ACID domain. The domain architecture consists of a closed β-barrel with seven strands (Β1-Β7) and three α-helices (H1-H3), an architecture showing similarities to that of the SPOC (Spen paralog and ortholog C-terminal domain) domain-like superfamily. Preliminary NMR chemical shift mapping showed that VP16 H2 (VP16C) interacts with MED25 ACID through one face of the β-barrel, defined by strands B4-B7-B6.
- CNRS UPR 2301, Chimie et biochimie structurales, Institut de Chimie des Substances Naturelles, Centre de recherche de Gif-sur-Yvette, 91190 Gif-sur-Yvette, France.
Organizational Affiliation: