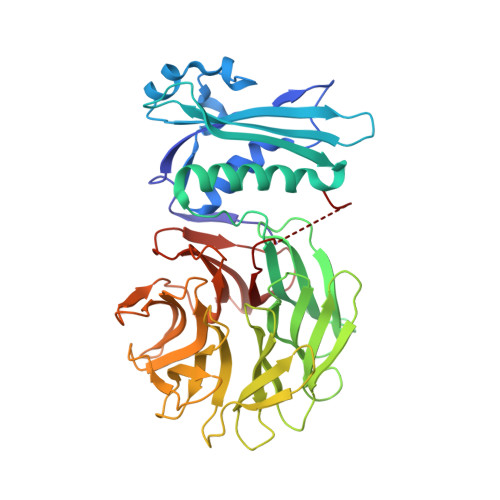
Molecular mimicry enables competitive recruitment by a natively disordered protein.
Bonsor, D.A., Grishkovskaya, I., Dodson, E.J., Kleanthous, C.(2007) J Am Chem Soc 129: 4800-4807
- PubMed: 17375930
- DOI: https://doi.org/10.1021/ja070153n
- Primary Citation of Related Structures:
2HQS - PubMed Abstract:
We report the crystal structure of the Escherichia coli TolB-Pal complex, a protein-protein complex involved in maintaining the integrity of the outer membrane (OM) in all Gram-negative bacteria that is parasitized by colicins (protein antibiotics) to expedite their entry into cells. Nuclease colicins competitively recruit TolB using their natively disordered regions (NDRs) to disrupt its complex with Pal, which is thought to trigger translocation of the toxin across a locally destabilized OM. The structure shows induced-fit binding of peptidoglycan-associated lipoprotein (Pal) to the beta-propeller domain of TolB causing the N-terminus of one of its alpha-helices to unwind and several residues to undergo substantial changes in conformation. The resulting interactions with TolB are known to be essential for the stability of the complex and the bacterial OM. Structural comparisons with a TolB-colicin NDR complex reveal that colicins bind at the Pal site, mimicking rearranged Pal residues while simultaneously appearing to block induced-fit changes in TolB. The study therefore explains how colicins recruit TolB in the bacterial periplasm and highlights a novel binding mechanism for a natively disordered protein.
- Department of Biology, University of York, Heslington, York, YO10 5YW, United Kingdom.
Organizational Affiliation: