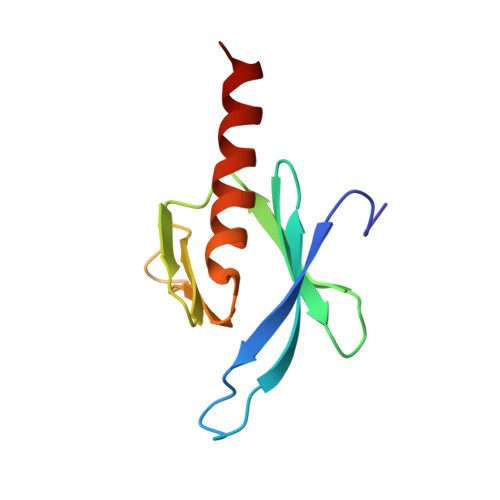
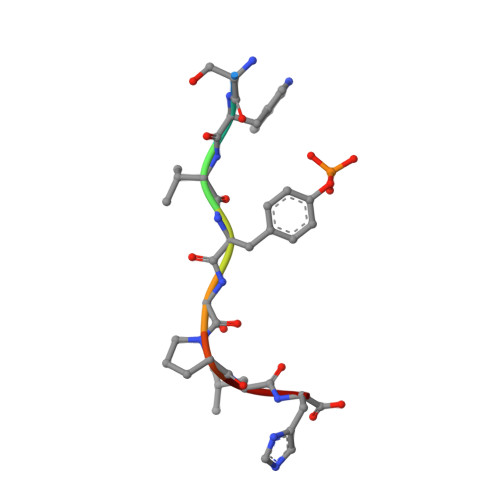
Structural Basis for the Phosphorylation-regulated Focal Adhesion Targeting of Type Igamma Phosphatidylinositol Phosphate Kinase (PIPKIgamma) by Talin.
Kong, X., Wang, X., Misra, S., Qin, J.(2006) J Mol Biology 359: 47-54
- PubMed: 16616931
- DOI: https://doi.org/10.1016/j.jmb.2006.02.048
- Primary Citation of Related Structures:
2G35 - PubMed Abstract:
Phosphatidylinositol-4,5-bisphosphate (PIP2) is a key lipid messenger that regulates myriad diverse cellular signaling pathways. To ensure specificity in disparate cellular events, PIP2 must be localized to specific sub-cellular sites. At PIP2-regulated focal adhesion (FA) sites, such localization is in part mediated via the recruitment and activation of PIP2-producing enzyme, type Igamma phosphatidylinositol phosphate kinase (PIPKIgamma), by a phosphotyrosine binding (PTB) domain of talin. Transient phosphorylation of PIPKIgamma at Y644 regulates the interaction and efficient FA targeting of PIPKIgamma; however, the underlying structural basis remains elusive. We have determined the NMR structure of talin-1 PTB in complex with the Y644-phosphorylated PIPKIgamma fragment (WVpYSPLH). As compared to canonical PTB domains that typically recognize the NPXpY turn motif from a variety of signaling proteins, our structure displays an unusual non-NPXpY-based recognition mode for talin-1 PTB where K(357)RW in beta5 strand forms an antiparallel beta-sheet with the VpYS of PIPKIgamma. A specific electrostatic triad between K357/R358 of talin-1 PTB and the pY644 of PIPKIgamma was observed, which is consistent with the mutagenesis and isothermal calorimetry data. Combined with previous in vivo data, our results provide a framework for understanding how phosphorylation of Y644 in PIPKIgamma promotes its specific interaction with talin-1, leading to efficient local synthesis of PIP2 and dynamic regulation of integrin-mediated FA assembly.
- Structural Biology Program, Lerner Research Institute, The Cleveland Clinic Foundation, Cleveland State University, OH 44195, USA.
Organizational Affiliation: