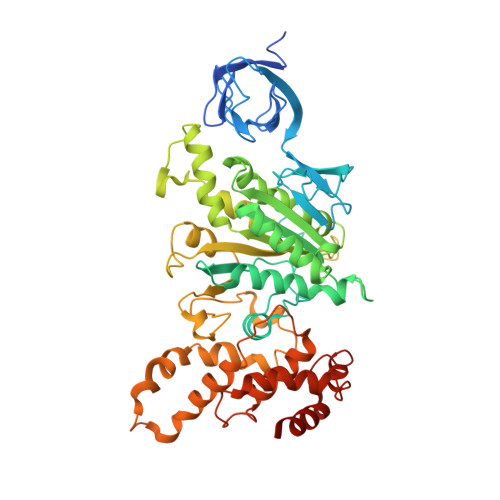
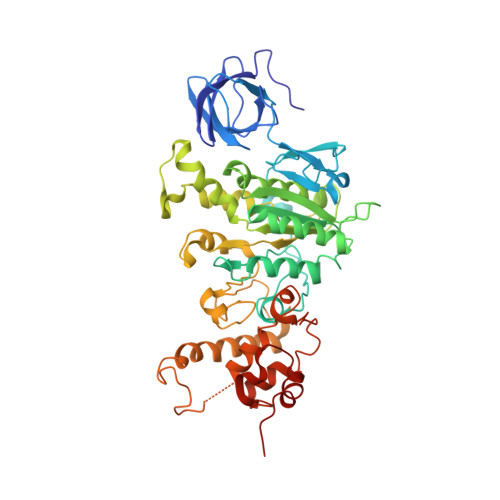
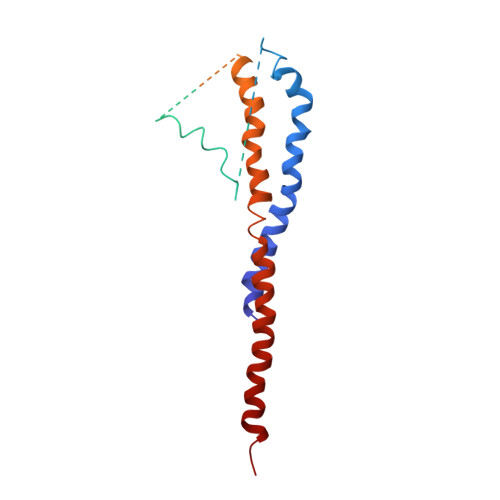
Mitochondrial ATP synthase: Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism.
Chen, C., Saxena, A.K., Simcoke, W.N., Garboczi, D.N., Pedersen, P.L., Ko, Y.H.(2006) J Biological Chem 281: 13777-13783
- PubMed: 16531409
- DOI: https://doi.org/10.1074/jbc.M513369200
- Primary Citation of Related Structures:
2F43 - PubMed Abstract:
ATP synthesis from ADP, P(i), and Mg2+ takes place in mitochondria on the catalytic F1 unit (alpha3beta3gammedeltaepsilon) of the ATP synthase complex (F0F1), a remarkable nanomachine that interconverts electrochemical and mechanical energy, producing the high energy terminal bond of ATP. In currently available structural models of F1, the P-loop (amino acid residues 156GGAGVGKT163) contributes to substrate binding at the subunit catalytic sites. Here, we report the first transition state-like structure of F1 (ADP.V(i).Mg.F1) from rat liver that was crystallized with the phosphate (P(i)) analog vanadate (VO(3-)4 or V(i)). Compared with earlier "ground state" structures, this new F1 structure reveals that the active site region has undergone significant remodeling. P-loop residue alanine 158 is located much closer to V(i) than it is to P(i) in a previous structural model. No significant movements of P-loop residues of the subunit were observed at its analogous but noncatalytic sites. Under physiological conditions, such active site remodeling involving the small hydrophobic alanine residue may promote ATP synthesis by lowering the local dielectric constant, thus facilitating the dehydration of ADP and P(i). This new crystallographic study provides strong support for the catalytic mechanism of ATP synthesis deduced from earlier biochemical studies of liver F1 conducted in the presence of V(i) (Ko, Y. H., Bianchet, M., Amzel, L. M., and Pedersen, P. L. (1997) J. Biol. Chem. 272, 18875-18881; Ko, Y. H., Hong, S., and Pedersen, P. L. (1999) J. Biol. Chem. 274, 28853-28856).
- Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205-2185.
Organizational Affiliation: