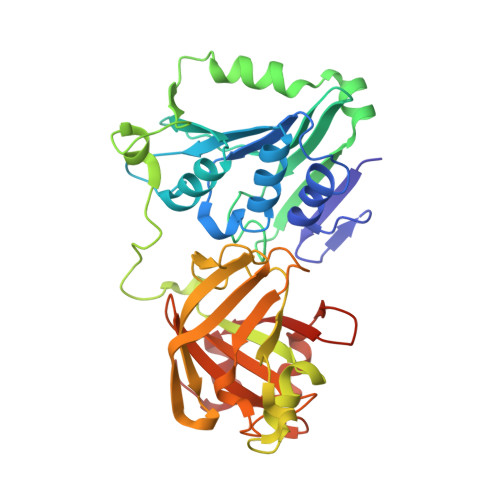
Structure of the Metal-Independent Restriction Enzyme Bfii Reveals Fusion of a Specific DNA-Binding Domain with a Nonspecific Nuclease.
Grazulis, S., Manakova, E., Roessle, M., Bochtler, M., Tamulaitiene, G., Huber, R., Siksnys, V.(2005) Proc Natl Acad Sci U S A 102: 15797
- PubMed: 16247004
- DOI: https://doi.org/10.1073/pnas.0507949102
- Primary Citation of Related Structures:
2C1L - PubMed Abstract:
Among all restriction endonucleases known to date, BfiI is unique in cleaving DNA in the absence of metal ions. BfiI represents a different evolutionary lineage of restriction enzymes, as shown by its crystal structure at 1.9-A resolution. The protein consists of two structural domains. The N-terminal catalytic domain is similar to Nuc, an EDTA-resistant nuclease from the phospholipase D superfamily. The C-terminal DNA-binding domain of BfiI exhibits a beta-barrel-like structure very similar to the effector DNA-binding domain of the Mg(2+)-dependent restriction enzyme EcoRII and to the B3-like DNA-binding domain of plant transcription factors. BfiI presumably evolved through domain fusion of a DNA-recognition element to a nonspecific nuclease akin to Nuc and elaborated a mechanism to limit DNA cleavage to a single double-strand break near the specific recognition sequence. The crystal structure suggests that the interdomain linker may act as an autoinhibitor controlling BfiI catalytic activity in the absence of a specific DNA sequence. A psi-blast search identified a BfiI homologue in a Mesorhizobium sp. BNC1 bacteria strain, a plant symbiont isolated from an EDTA-rich environment.
- Laboratory of Protein-DNA Interaction, Institute of Biotechnology, Graiciuno 8, LT-02241 Vilnius, Lithuania. grazulis@ibt.lt
Organizational Affiliation: