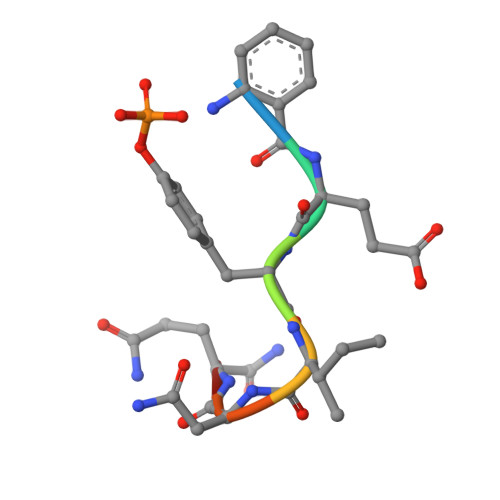
Structural basis for the high affinity of amino-aromatic SH2 phosphopeptide ligands.
Rahuel, J., Garcia-Echeverria, C., Furet, P., Strauss, A., Caravatti, G., Fretz, H., Schoepfer, J., Gay, B.(1998) J Mol Biology 279: 1013-1022
- PubMed: 9642078
- DOI: https://doi.org/10.1006/jmbi.1998.1790
- Primary Citation of Related Structures:
1ZFP - PubMed Abstract:
An anthranyl moiety placed at the N terminus of a phosphotyrosine peptide potentiates the inhibitory effect of this small peptide on the binding of the Grb2 SH2 domain to the EGF receptor. Using molecular modeling procedures based on the Lck SH2 domain structure, this observation was rationalized in terms of a suitably favorable pi-pi stacking interaction between the anthranyl moiety and the arginine alphaA2 (ArgalphaA2) residue side-chain of Grb2 SH2. The crystal structure of the Grb2 SH2 domain in complex with the inhibitor 2-Abz-EpYINQ-NH2 (IC50 26 nM) has been solved in two different crystal forms at 2.1 and 1.8 A resolution. This structure confirms the modeling based on the Lck SH2 domain. The ArgalphaA2 residue is conserved in most SH2 domains. Thus, as expected, the anthranyl group also confers high affinity to small peptide ligands of other SH2 domains such as Lck-, PLC-gamma-amino-terminal and p85 amino-terminal SH2 domains as demonstrated by structure affinity relationships (SAR) data. These potent peptides with an amino-terminal surrogate group and the structure of Grb2 SH2 domain in complex with one such peptide represent good starting points for the design and optimization of new inhibitors of many SH2 domains.
- Core Technology Area, Novartis Pharma AG, Core Technology Area and Oncology, Research Department, CH-4002, Basel Switzerland.
Organizational Affiliation: