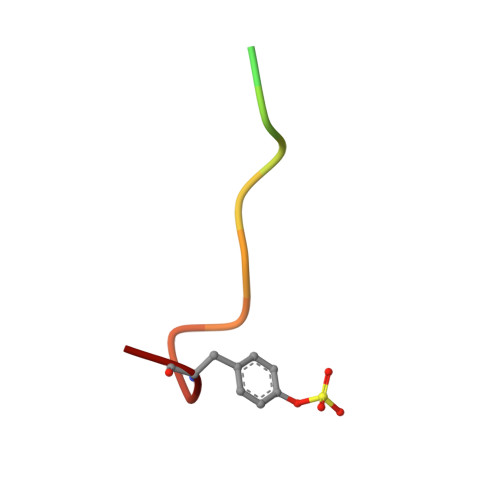
Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Krishnan, R., Mochalkin, I., Arni, R., Tulinsky, A.(2000) Acta Crystallogr D Biol Crystallogr 56: 294-303
- PubMed: 10713516
- DOI: https://doi.org/10.1107/s0907444900000068
- Primary Citation of Related Structures:
1C4U, 1C4V, 1C4Y, 1D6W, 1D9I - PubMed Abstract:
The crystal structures of five new non-electrophilic beta-strand-templated thrombin active-site inhibitors have been determined bound to the enzyme. Four co-crystallize with hirugen and inhibitor isomorphously to produce thrombin-hirugen crystals (monoclinic, space group C2), while one co-crystallizes in the hexagonal system, space group P6(5). A 1,4-substituted cyclohexyl moiety is conserved at the P1 position of all the inhibitors, along with a fused hetero-bicyclic five- and six-membered ring that occupies the P2 site. Amino, amidino and aminoimidazole groups are attached to the cyclohexyl ring for recognition at the S1 specificity site, while benzylsulfonyl and diphenyl groups enhance the binding at the S3 subsite. The cyclohexyl groups at the P1 positions of three of the inhibitors appear to be in the energetically favored chair conformation, while the imidazole-substituted cyclohexyl rings are in a boat conformation. Somewhat unexpectedly, the two cyclohexyl-aminoimidazole groups bind differently in the specificity site; the unique binding of one is heretofore unreported. The other inhibitors generally mimic arginyl binding at S1. This group of inhibitors combines the non-electrophilicity and selectivity of DAPA-like compounds and the more optimal binding features of the S1-S3 sites of thrombin for peptidic molecules, which results in highly potent (binding constants 12 nM-16 pM, one being 1.1 microM) and selective (ranging from 140 to 20 000 times more selective compared with trypsin) inhibitors of thrombin. The binding modes of these novel inhibitors are correlated with their binding constants, as is their selectivity, in order to provide further insight for the design of therapeutic antithrombotic agents that inhibit thrombin directly at the active site.
- Department of Chemistry, Michigan State University, East Lansing, Michigan 48824-1322, USA.
Organizational Affiliation: