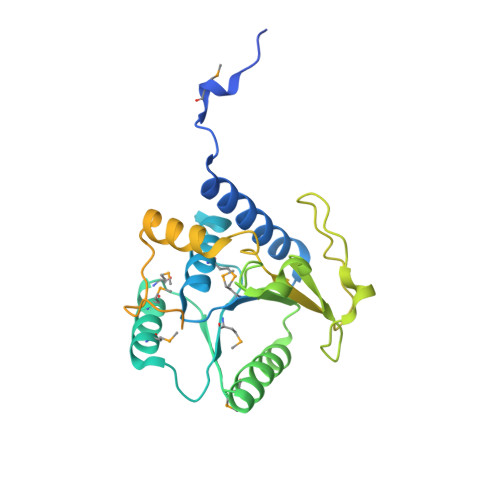
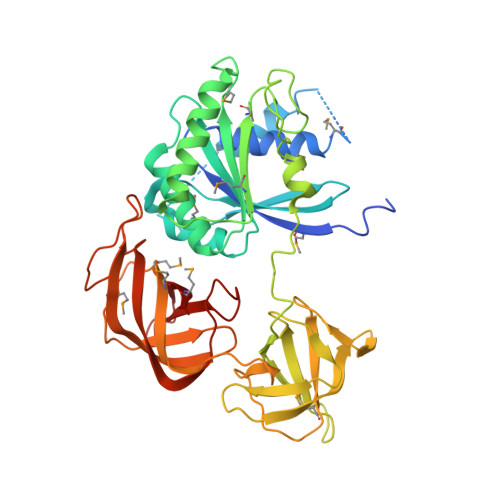
Molecular basis for g protein control of the prokaryotic ATP sulfurylase.
Mougous, J.D., Lee, D.H., Hubbard, S.C., Schelle, M.W., Vocadlo, D.J., Berger, J.M., Bertozzi, C.R.(2006) Mol Cell 21: 109-122
- PubMed: 16387658
- DOI: https://doi.org/10.1016/j.molcel.2005.10.034
- Primary Citation of Related Structures:
1ZUN - PubMed Abstract:
Sulfate assimilation is a critical component of both primary and secondary metabolism. An essential step in this pathway is the activation of sulfate through adenylation by the enzyme ATP sulfurylase (ATPS), forming adenosine 5'-phosphosulfate (APS). Proteobacterial ATPS overcomes this energetically unfavorable reaction by associating with a regulatory G protein, coupling the energy of GTP hydrolysis to APS formation. To discover the molecular basis of this unusual role for a G protein, we biochemically characterized and solved the X-ray crystal structure of a complex between Pseudomonas syringae ATPS (CysD) and its associated regulatory G protein (CysN). The structure of CysN*D shows the two proteins in tight association; however, the nucleotides bound to each subunit are spatially segregated. We provide evidence that conserved switch motifs in the G domain of CysN allosterically mediate interactions between the nucleotide binding sites. This structure suggests a molecular mechanism by which conserved G domain architecture is used to energetically link GTP turnover to the production of an essential metabolite.
- Howard Hughes Medical Institute, University of California, Berkeley, Berkeley, California 94720, USA.
Organizational Affiliation: