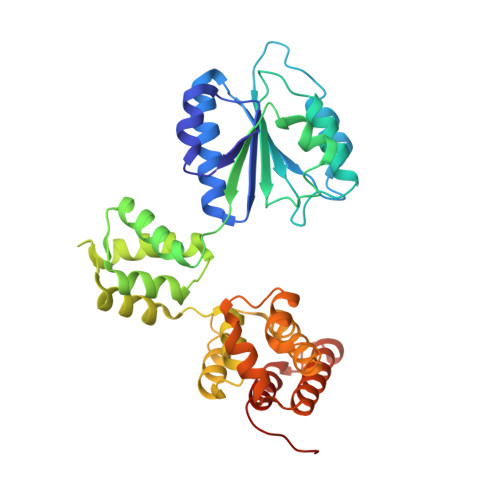
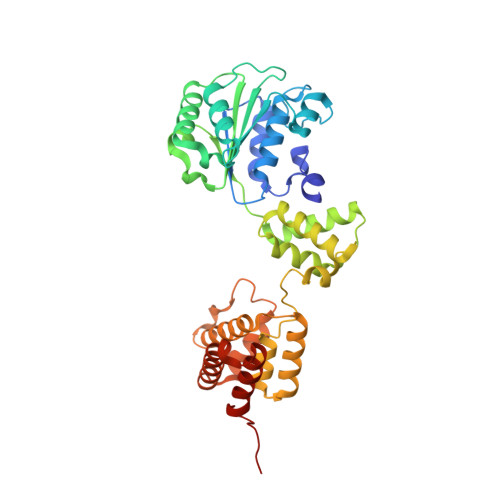
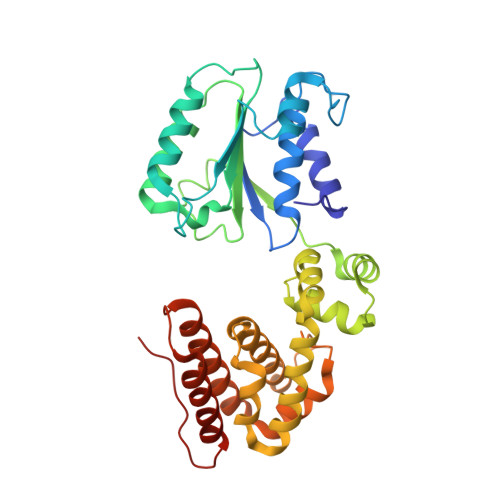
Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Kazmirski, S.L., Podobnik, M., Weitze, T.F., O'donnell, M., Kuriyan, J.(2004) Proc Natl Acad Sci U S A 101: 16750-16755
- PubMed: 15556993
- DOI: https://doi.org/10.1073/pnas.0407904101
- Primary Citation of Related Structures:
1XXH, 1XXI - PubMed Abstract:
Clamp-loader complexes are heteropentameric AAA+ ATPases that load sliding clamps onto DNA. The structure of the nucleotide-free Escherichia coli clamp loader had been determined previously and led to the proposal that the clamp-loader cycles between an inactive state, in which the ATPase domains form a closed ring, and an active state that opens up to form a "C" shape. The crystal structure was interpreted as being closer to the active state than the inactive state. The crystal structure of a nucleotide-bound eukaryotic clamp loader [replication factor C (RFC)] revealed a different and more tightly packed spiral organization of the ATPase domains, raising questions about the significance of the conformation seen earlier for the bacterial clamp loader. We describe crystal structures of the E. coli clamp-loader complex bound to the ATP analog ATPgammaS (at a resolution of 3.5 A) and ADP (at a resolution of 4.1 A). These structures are similar to that of the nucleotide-free clamp-loader complex. Only two of the three functional ATP-binding sites are occupied by ATPgammaS or ADP in these structures, and the bound nucleotides make no interfacial contacts in the complex. These results, along with data from isothermal titration calorimetry, molecular dynamics simulations, and comparison with the RFC structure, suggest that the more open form of the E. coli clamp loader described earlier and in the present work corresponds to a stable inactive state of the clamp loader in which the ATPase domains are prevented from engaging the clamp in the highly cooperative manner seen in the fully ATP-loaded RFC-clamp structure.
- Department of Molecular and Cell Biology, Howard Hughes Medical Institute, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: