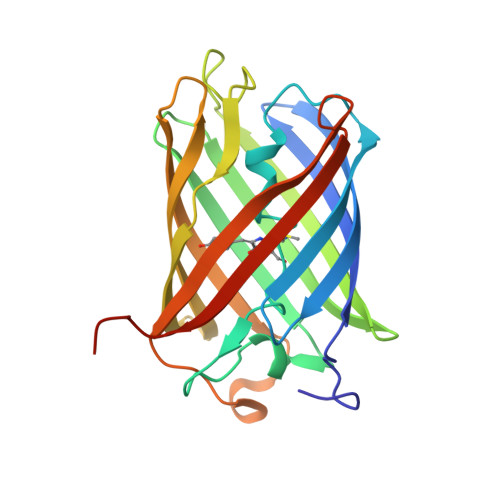
The 2.0A crystal structure of eqFP611, a far-red fluorescent protein from the sea anemone Entacmaea quadricolor
Petersen, J., Wilmann, P.G., Beddoe, T., Oakley, A.J., Devenish, R.J., Prescott, M., Rossjohn, J.(2003) J Biological Chem 278: 44626-44631
- PubMed: 12909624
- DOI: https://doi.org/10.1074/jbc.M307896200
- Primary Citation of Related Structures:
1UIS - PubMed Abstract:
We have crystallized and subsequently determined to 2.0-A resolution the crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. The structure of the protomer, which adopts a beta-can topology, is similar to that of the related monomeric green fluorescent protein (GFP). The quaternary structure of eqFP611, a tetramer exhibiting 222 symmetry, is similar to that observed for the more closely related red fluorescent protein DsRed and the chromoprotein Rtms5. The unique chromophore sequence (Met63-Tyr64-Gly65) of eqFP611, adopts a coplanar and trans conformation within the interior of the beta-can fold. Accordingly, the eqFP611 chromophore adopts a significantly different conformation in comparison to the chromophore conformation observed in GFP, DsRed, and Rtms5. The coplanar chromophore conformation and its immediate environment provide a structural basis for the far red, highly fluorescent nature of eqFP611. The eqFP611 structure extends our knowledge on the range of conformations a chromophore can adopt within closely related members of the green fluorescent protein family.
- The Protein Crystallography Unit, School of Biomedical Sciences, Monash University, Victoria, Australia.
Organizational Affiliation: