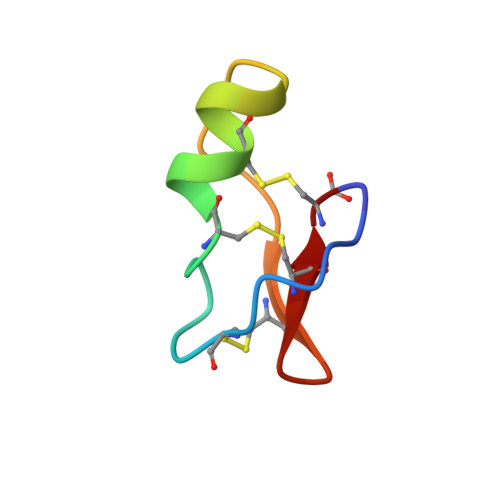
Solution structure of TsKapa, a charybdotoxin-like scorpion toxin from Tityus serrulatus with high affinity for apamin-sensitive Ca(2+)-activated K+ channels.
Blanc, E., Lecomte, C., Rietschoten, J.V., Sabatier, J.M., Darbon, H.(1997) Proteins 29: 359-369
- PubMed: 9365990
- DOI: https://doi.org/10.1002/(sici)1097-0134(199711)29:3<359::aid-prot9>3.0.co;2-5
- Primary Citation of Related Structures:
1TSK - PubMed Abstract:
TsKapa (TsK), purified from the Buthidae Tityus serrulatus is a very high potent ligand for small-conductance apamin-sensitive calcium-activated potassium channels (SK). It is able to efficiently compete with apamin for binding on this channel (K0.5 = 0.3 nM) [Legros, C. et al., FEBS Lett. 390:81-84, 1996]. The solution structure of TsK has been determined by 2D-NMR techniques, which led to the full description of its 3D conformation: a short alpha helix from residues 14 to 20 and a three-stranded antiparallel beta sheet (residues 2-3, 27-29, and 32-34). The interaction of TsK with the SK potassium channel has been modeled according to the charge anisotropy of the ligand. The resulting dipole moment orientates TsK so that it presents toward the receptor, a surface, mainly basic, encompassing residues K18 and K19 on one side and R9 and Y8 on the other. Despite its three-dimensional structure that is related with scorpion toxins active on voltage-gated potassium channels such as charybdotoxin, the pharmacological activity and specificity of TsK is related with shorter scorpion toxins (i.e., possessing an only two-stranded beta sheet) such as scyllatoxin (also named leiurotoxin I) or P05.
- AFMB, CNRS UPR 9039, Marseille, France.
Organizational Affiliation: