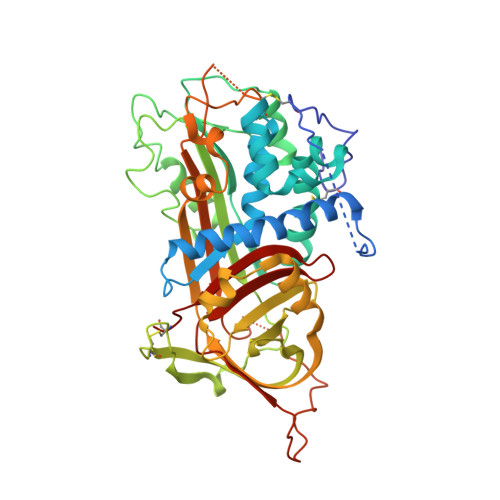
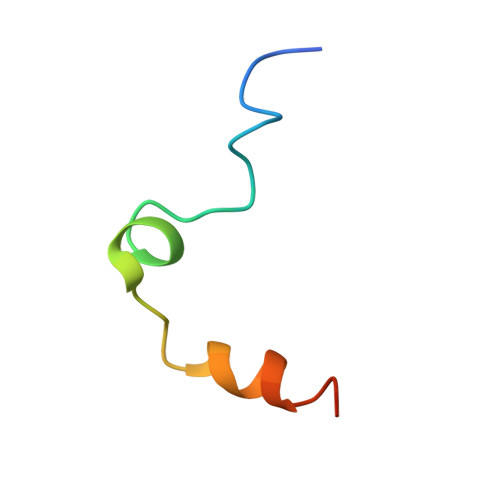
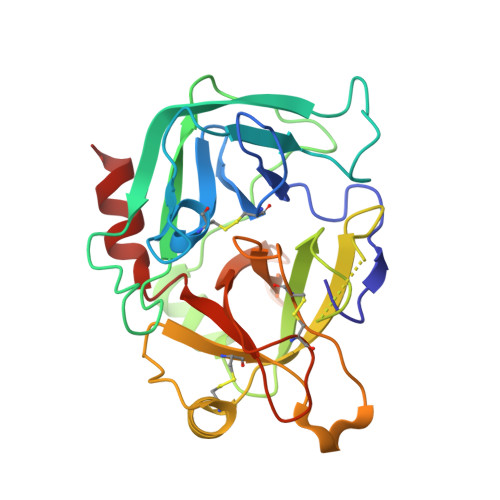
The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity.
Dementiev, A., Petitou, M., Herbert, J.M., Gettins, P.G.(2004) Nat Struct Mol Biol 11: 863-867
- PubMed: 15311268
- DOI: https://doi.org/10.1038/nsmb810
- Primary Citation of Related Structures:
1SR5 - PubMed Abstract:
Antithrombin, the principal physiological inhibitor of the blood coagulation proteinase thrombin, requires heparin as a cofactor. We report the crystal structure of the rate-determining encounter complex formed between antithrombin, anhydrothrombin and an optimal synthetic 16-mer oligosaccharide. The antithrombin reactive center loop projects from the serpin body and adopts a canonical conformation that makes extensive backbone and side chain contacts from P5 to P6' with thrombin's restrictive specificity pockets, including residues in the 60-loop. These contacts rationalize many earlier mutagenesis studies on thrombin specificity. The 16-mer oligosaccharide is just long enough to form the predicted bridge between the high-affinity pentasaccharide-binding site on antithrombin and the highly basic exosite 2 on thrombin, validating the design strategy for this synthetic heparin. The protein-protein and protein-oligosaccharide interactions together explain the basis for heparin activation of antithrombin as a thrombin inhibitor.
- Department of Biochemistry and Molecular Genetics, College of Medicine, University of Illinois at Chicago, 900 S. Ashland, Chicago, Illinois 60607, USA.
Organizational Affiliation: