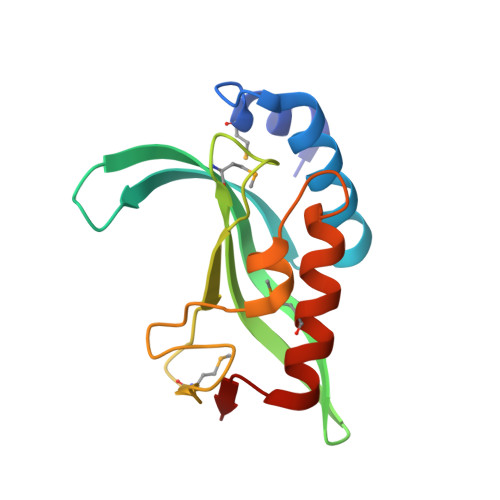
Ubiquitin recognition by the human TSG101 protein
Sundquist, W.I., Schubert, H.L., Kelly, B.N., Hill, G.C., Holton, J.M., Hill, C.P.(2004) Mol Cell 13: 783-789
- PubMed: 15053872
- DOI: https://doi.org/10.1016/s1097-2765(04)00129-7
- Primary Citation of Related Structures:
1S1Q - PubMed Abstract:
The UEV domain of the TSG101 protein functions in both HIV-1 budding and the vacuolar protein sorting (VPS) pathway, where it binds ubiquitylated proteins as they are sorted into vesicles that bud into late endosomal compartments called multivesicular bodies (MVBs). TSG101 UEV-ubiquitin interactions are therefore important for delivery of both substrates and hydrolytic enzymes to lysosomes, which receive proteins via fusion with MVBs. Here, we report the crystal structure of the TSG101 UEV domain in complex with ubiquitin at 2.0 A resolution. TSG101 UEV contacts the Ile44 surface and an adjacent loop of ubiquitin through a highly solvated interface. Mutations that disrupt the interface inhibit MVB sorting, and the structure also explains how the TSG101 UEV can independently bind its ubiquitin and Pro-Thr/Ser-Ala-Pro peptide ligands. Remarkably, comparison with mapping data from other UEV and related E2 proteins indicates that although the different E2/UEV domains share the same structure and have conserved ubiquitin binding activity, they bind through very different interfaces.
- Department of Biochemistry, University of Utah, Salt Lake City, UT 84132, USA. wes@biochem.utah.edu
Organizational Affiliation: