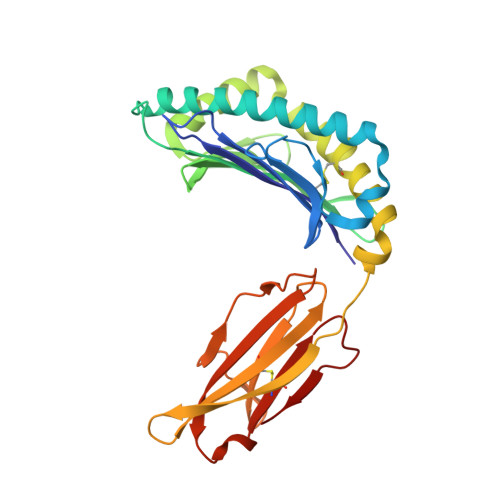
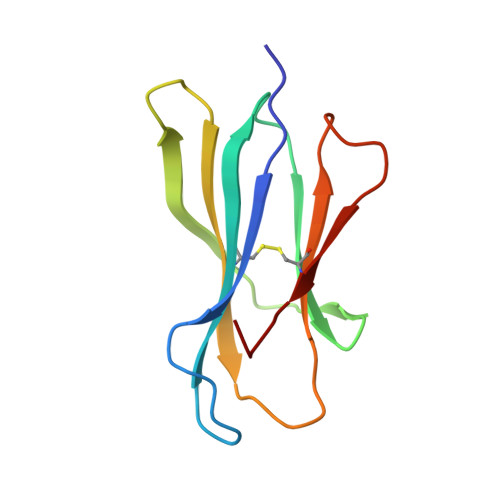
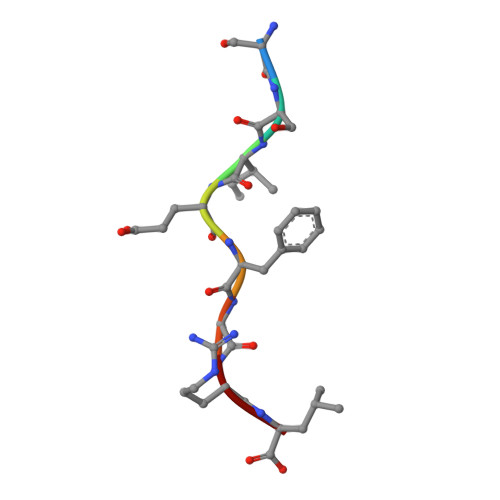
Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
Miley, M.J., Messaoudi, I., Metzner, B.M., Wu, Y., Nikolich-Zugich, J., Fremont, D.H.(2004) J Exp Medicine 200: 1445-1454
- PubMed: 15557346
- DOI: https://doi.org/10.1084/jem.20040217
- Primary Citation of Related Structures:
1RJY, 1RJZ, 1RK0, 1RK1 - PubMed Abstract:
Major histocompatibility complex (MHC) class I variants H-2K(b) and H-2K(bm8) differ primarily in the B pocket of the peptide-binding groove, which serves to sequester the P2 secondary anchor residue. This polymorphism determines resistance to lethal herpes simplex virus (HSV-1) infection by modulating T cell responses to the immunodominant glycoprotein B(498-505) epitope, HSV8. We studied the molecular basis of these effects and confirmed that T cell receptors raised against K(b)-HSV8 cannot recognize H-2K(bm8)-HSV8. However, substitution of Ser(P2) to Glu(P2) (peptide H2E) reversed T cell receptor (TCR) recognition; H-2K(bm8)-H2E was recognized whereas H-2K(b)-H2E was not. Insight into the structural basis of this discrimination was obtained by determining the crystal structures of all four MHC class I molecules in complex with bound peptide (pMHCs). Surprisingly, we find no concerted pMHC surface differences that can explain the differential TCR recognition. However, a correlation is apparent between the recognition data and the underlying peptide-binding groove chemistry of the B pocket, revealing that secondary anchor residues can profoundly affect TCR engagement through mechanisms distinct from the alteration of the resting state conformation of the pMHC surface.
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO 63110, USA.
Organizational Affiliation: