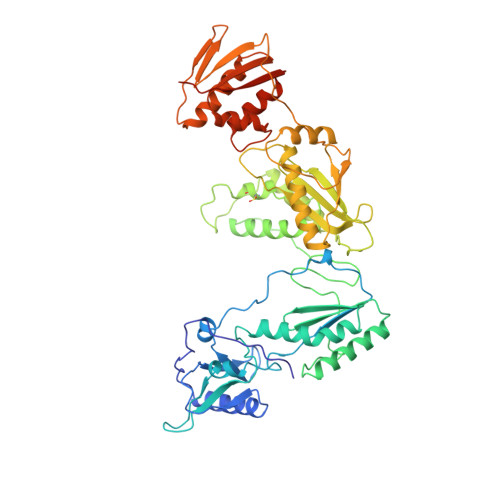
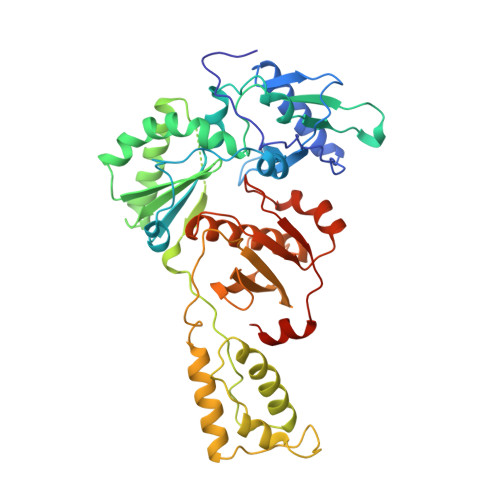
The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design.
Ren, J., Esnouf, R., Hopkins, A., Ross, C., Jones, Y., Stammers, D., Stuart, D.(1995) Structure 3: 915-926
- PubMed: 8535785
- DOI: https://doi.org/10.1016/S0969-2126(01)00226-X
- Primary Citation of Related Structures:
1REV - PubMed Abstract:
HIV reverse transcriptase (RT) is a key target of anti-AIDS therapies. Structural studies of HIV-1 RT, unliganded and complexed with different non-nucleoside inhibitors (NNIs), have pointed to a common mode of binding and inactivation through distortion of the polymerase catalytic site by NNIs containing two hinged rings. The mode of binding of the TIBO family of inhibitors is of interest because these compounds do not fit the two-hinged-ring model. The structure of HIV-1 RT complexed with 9-chloro-TIBO (R82913) has been determined at 2.6 A resolution. As reported for the lower resolution analysis of another TIBO compound, this inhibitor binds at the same site as other NNIs, but our higher resolution study reveals the Cl-TIBO is distorted from the conformation seen in crystals of the inhibitor alone. This allows Cl-TIBO to mimic the binding of NNIs containing two hinged rings. Inhibitor-protein interactions are again predominantly hydrophobic and the protein conformation corresponds to that seen in complexes with other tight-binding NNIs. Although Cl-TIBO is chemically very different from other NNIs, it achieves remarkable spatial equivalence and shape complementarity with other NNIs on binding to RT. Comparison of the different RT-NNI complexes suggests modifications to the TIBO group of inhibitors which might enhance their binding and hence, potentially, their therapeutic efficacy.
- Laboratory of Molecular Biophysics, Oxford, UK.
Organizational Affiliation: