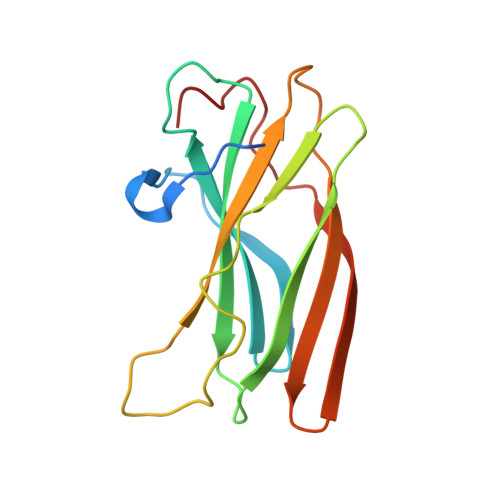
Structural basis for binding of accessory proteins by the appendage domain of GGAs
Collins, B.M., Praefcke, G.J.K., Robinson, M.S., Owen, D.J.(2003) Nat Struct Biol 10: 607-613
- PubMed: 12858163
- DOI: https://doi.org/10.1038/nsb955
- Primary Citation of Related Structures:
1OM9 - PubMed Abstract:
The Golgi-associated, gamma-adaptin-related, ADP-ribosylation-factor binding proteins (GGAs) and adaptor protein (AP)-1 are adaptors involved in clathrin-mediated transport between the trans-Golgi network and endosomal system. The appendage domains of GGAs and the AP-1 gamma-adaptin subunit are structurally homologous and have been proposed to bind to accessory proteins via interaction with short sequences containing phenylalanines and acidic residues. Here we present the structure of the human GGA1 appendage in complex with its cognate binding peptide from the p56 accessory protein (DDDDFGGFEAAETFD) as determined by X-ray crystallography. The interaction is governed predominantly by packing of the first two phenylalanine residues of the peptide with conserved basic and hydrophobic residues from GGA1. Additionally, several main chain hydrogen bonds cause the peptide to form an additional beta-strand on the edge of the preexisting beta-sheet of the protein. Isothermal titration calorimetry was used to assess the affinities of different peptides for the GGA and gamma-appendage domains.
- Cambridge Institute for Medical Research, Department of Clinical Biochemistry, University of Cambridge, Hills Road, Cambridge CB2 2XY, UK. bmc25@cam.ac.uk
Organizational Affiliation: