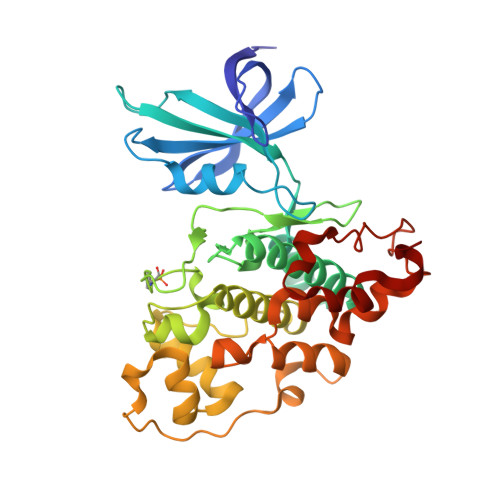
Structural Basis for Recruitment of Glycogen Synthase Kinase 3Beta to the Axin-Apc Scaffold Complex
Dajani, R., Fraser, E., Roe, S.M., Yeo, M., Good, V., Thompson, V., Dale, T.C., Pearl, L.H.(2003) EMBO J 22: 494
- PubMed: 12554650
- DOI: https://doi.org/10.1093/emboj/cdg068
- Primary Citation of Related Structures:
1O9U - PubMed Abstract:
Glycogen synthase kinase 3beta (GSK3beta) is a serine/threonine kinase involved in insulin, growth factor and Wnt signalling. In Wnt signalling, GSK3beta is recruited to a multiprotein complex via interaction with axin, where it hyperphosphorylates beta-catenin, marking it for ubiquitylation and destruction. We have now determined the crystal structure of GSK3beta in complex with a minimal GSK3beta-binding segment of axin, at 2.4 A resolution. The structure confirms the co-localization of the binding sites for axin and FRAT in the C-terminal domain of GSK3beta, but reveals significant differences in the interactions made by axin and FRAT, mediated by conformational plasticity of the 285-299 loop in GSK3beta. Detailed comparison of the axin and FRAT GSK3beta complexes allows the generation of highly specific mutations, which abrogate binding of one or the other. Quantitative analysis suggests that the interaction of GSK3beta with the axin scaffold enhances phosphorylation of beta-catenin by >20 000-fold.
- Section of Structural Biology and Cancer Research UK Centre for Cell and Molecular Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, UK.
Organizational Affiliation: