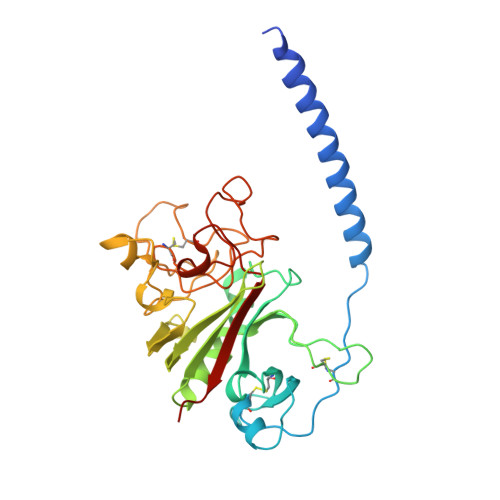
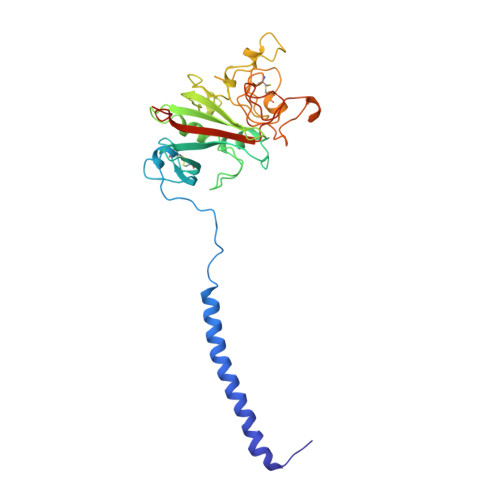
2.8 A Crystal Structures of Recombinant Fibrinogen Fragment D with and without Two Peptide Ligands: GHRP Binding to the "b" Site Disrupts Its Nearby Calcium-binding Site.
Kostelansky, M.S., Betts, L., Gorkun, O.V., Lord, S.T.(2002) Biochemistry 41: 12124-12132
- PubMed: 12356313
- DOI: https://doi.org/10.1021/bi0261894
- Primary Citation of Related Structures:
1LT9, 1LTJ - PubMed Abstract:
We report two crystal structures, each at a resolution of 2.8 A, of recombinant human fibrinogen fragment D (rfD) in the absence and presence of peptide ligands. The bound ligands, Gly-Pro-Arg-Pro-amide and Gly-His-Arg-Pro-amide, mimic the interactions of the thrombin exposed polymerization sites, "A" and "B", respectively. This report is the first to describe the structure of fragment D in the presence of both peptide ligands. The structures reveal that recombinant fibrinogen is nearly identical to the plasma protein but with minor changes, like the addition of a proximal fucose to the carbohydrate linked to residue betaGln364, and slightly different relative positions of the beta- and gamma-modules. Of major interest in our structures is that a previously identified calcium site in plasma fibrinogen is absent when Gly-His-Arg-Pro-amide is bound. The peptide-dependent loss of this calcium site may have significant biological implications that are further discussed. These structures provide a foundation for the detailed structural analysis of variant recombinant fibrinogens that were used to identify critical functional residues within fragment D.
- Department of Chemistry, University of North Carolina, Chapel Hill, NC 27599, USA.
Organizational Affiliation: