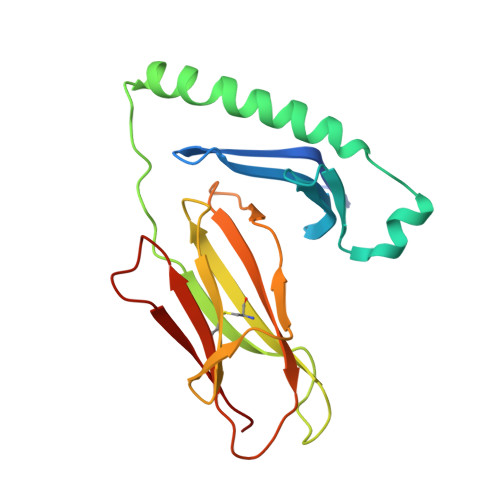
Structural snapshot of aberrant antigen presentation linked to autoimmunity: the immunodominant epitope of MBP complexed with I-Au
He, X.L., Radu, C., Sidney, J., Sette, A., Ward, E.S., Garcia, K.C.(2002) Immunity 17: 83-94
- PubMed: 12150894
- DOI: https://doi.org/10.1016/s1074-7613(02)00340-0
- Primary Citation of Related Structures:
1K2D - PubMed Abstract:
Murine experimental allergic encephalomyelitis (EAE) is a useful model for the demyelinating, autoimmune disease multiple sclerosis. In the EAE system, the immunodominant N-terminal epitope of myelin basic protein (MBP) is an unusually short, weakly binding peptide antigen which elicits highly biased TCR chain usage. In the 2.2 A crystal structure of I-A(u)/MBP1-11 complex, only MBP residues 1-7 are bound toward one end of the peptide binding cleft. The fourth residue of MBP1-11 is located in an incompatible p6 pocket of I-A(u), thus explaining the short half-life of I-A(u) complexed with Ac1-11. MBP peptides extended at the C terminus of Ac1-11 result in dramatic affinity increases, likely attributed to register shifting to a higher affinity cryptic epitope, which could potentially mask the presentation of the immunodominant MBP1-11 peptide during thymic education.
- Department of Microbiology and Immunology, Stanford University School of Medicine, CA 94305, USA.
Organizational Affiliation: